Abstract
Purpose
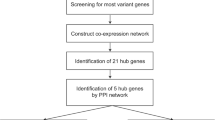
Breast cancer is one of the most common malignancies among females, and its prognosis is affected by a complex network of gene interactions. Weighted gene co-expression network analysis was used to construct free-scale gene co-expression networks and to identify potential biomarkers for breast cancer progression.
Methods
The gene expression profiles of GSE42568 were downloaded from the Gene Expression Omnibus database. RNA-sequencing data and clinical information of breast cancer from TCGA were used for validation.
Results
A total of ten modules were established by the average linkage hierarchical clustering. We identified 58 network hub genes in the significant module (R2 = 0.44) and 6 hub genes (AGO2, CDC20, CDCA5, MCM10, MYBL2, and TTK), which were significantly correlated with prognosis. Receiver-operating characteristic curve validated that the mRNA levels of these six genes exhibited excellent diagnostic efficiency in the test data set of GSE42568. RNA-sequencing data from TCGA showed that the expression levels of these six genes were higher in triple-negative tumors. One-way ANOVA suggested that these six genes were upregulated at more advanced stages. The results of independent sample t test indicated that MCM10 and TTK were associated with tumor size, and that AGO2, CDC20, CDCA5, MCM10, and MYBL2 were overexpressed in lymph-node positive breast cancer.
Conclusions
AGO2, CDC20, CDCA5, MCM10, MYBL2, and TTK were identified as candidate biomarkers for further basic and clinical research on breast cancer based on co-expression analysis.








Similar content being viewed by others
References
Adams BD, Claffey KP, White BA (2009) Argonaute-2 expression is regulated by epidermal growth factor receptor and mitogen-activated protein kinase signaling and correlates with a transformed phenotype in breast cancer cells. Endocrinology 150:14–23. https://doi.org/10.1210/en.2008-0984
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 68:394–424. https://doi.org/10.3322/caac.21492
Chang IW et al (2015) CDCA5 overexpression is an indicator of poor prognosis in patients with urothelial carcinomas of the upper urinary tract and urinary bladder. Am J Transl Res 7:710–722
Chen T et al (2017) Role of triosephosphate isomerase and downstream functional genes on gastric cancer. Oncol Rep 38:1822–1832. https://doi.org/10.3892/or.2017.5846
Chen X et al (2018) A novel USP9X substrate TTK contributes to tumorigenesis in non-small-cell lung cancer. Theranostics 8:2348–2360. https://doi.org/10.7150/thno.22901
da Huang W, Sherman BT, Lempicki RA (2009a) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37:1–13. https://doi.org/10.1093/nar/gkn923
da Huang W, Sherman BT, Lempicki RA (2009b) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. https://doi.org/10.1038/nprot.2008.211
Fan X et al (2018) B-Myb mediates proliferation and migration of non-small-cell lung cancer via suppressing IGFBP3. Int J Mol Sci. https://doi.org/10.3390/ijms19051479
Fisher B, Land S, Mamounas E, Dignam J, Fisher ER, Wolmark N (2001) Prevention of invasive breast cancer in women with ductal carcinoma in situ: an update of the National Surgical Adjuvant Breast and Bowel Project experience. Semin Oncol 28:400–418
Ford D, Easton DF, Bishop DT, Narod SA, Goldgar DE (1994) Risks of cancer in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Lancet (Lond, Engl) 343:692–695
Gao Y, Zhang B, Wang Y, Shang G (2018) Cdc20 inhibitor apcin inhibits the growth and invasion of osteosarcoma cells. Oncol Rep 40:841–848. https://doi.org/10.3892/or.2018.6467
Gonzalez-Angulo AM et al (2011) Incidence and outcome of BRCA mutations in unselected patients with triple receptor-negative breast cancer. Clin Cancer Res 17:1082–1089. https://doi.org/10.1158/1078-0432.Ccr-10-2560
Guan Z, Cheng W, Huang D, Wei A (2018) High MYBL2 expression and transcription regulatory activity is associated with poor overall survival in patients with hepatocellular carcinoma. Curr Res Transl Med 66:27–32. https://doi.org/10.1016/j.retram.2017.11.002
Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z (2010) An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res Treat 123:725–731. https://doi.org/10.1007/s10549-009-0674-9
Jung KH et al (2018) Adjuvant subcutaneous trastuzumab for HER2-positive early breast cancer: subgroup analyses of safety and active medical conditions by body weight in the SafeHer phase III study. The oncologist 23:1137–1143. https://doi.org/10.1634/theoncologist.2018-0065
Kang G et al (2013) Exome sequencing identifies early gastric carcinoma as an early stage of advanced gastric cancer. PloS One 8:e82770. https://doi.org/10.1371/journal.pone.0082770
Khalil MY, Grandis JR, Shin DM (2003) Targeting epidermal growth factor receptor: novel therapeutics in the management of cancer. Expert Rev Anticancer Ther 3:367–380. https://doi.org/10.1586/14737140.3.3.367
King CR, Kraus MH, Aaronson SA (1985) Amplification of a novel v-erbB-related gene in a human mammary carcinoma. Science (New York) 229:974–976
Landi MT et al (2008) Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PloS One 3:e1651. https://doi.org/10.1371/journal.pone.0001651
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinform 9:559. https://doi.org/10.1186/1471-2105-9-559
Li WM et al (2016) MCM10 overexpression implicates adverse prognosis in urothelial carcinoma. Oncotarget 7:77777–77792. https://doi.org/10.18632/oncotarget.12795
Lin D et al (1994) Constitutive expression of B-myb can bypass p53-induced Waf1/Cip1-mediated G1 arrest. Proc Natl Acad Sci USA 91:10079–10083
Lu P et al (2014) Genome-wide gene expression profile analyses identify CTTN as a potential prognostic marker in esophageal cancer. PloS One 9:e88918. https://doi.org/10.1371/journal.pone.0088918
Mahadevappa R et al (2018) DNA replication licensing protein MCM10 promotes tumor progression and is a novel prognostic biomarker and potential therapeutic target in breast cancer. Cancers. https://doi.org/10.3390/cancers10090282
Mannefeld M, Klassen E, Gaubatz S (2009) B-MYB is required for recovery from the DNA damage-induced G2 checkpoint in p53 mutant cells. Cancer Res 69:4073–4080. https://doi.org/10.1158/0008-5472.can-08-4156
Musa J, Aynaud MM, Mirabeau O, Delattre O, Grunewald TG (2017) MYBL2 (B-Myb): a central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death Dis 8:e2895. https://doi.org/10.1038/cddis.2017.244
O’Shaughnessy J (2005) Extending survival with chemotherapy in metastatic breast cancer. Oncologist 10(Suppl 3):20–29. https://doi.org/10.1634/theoncologist.10-90003-20
Paterson MC et al (1991) Correlation between c-erbB-2 amplification and risk of recurrent disease in node-negative breast cancer. Cancer Res 51:556–567
Paul D, Ghorai S, Dinesh US, Shetty P, Chattopadhyay S, Santra MK (2017) Cdc20 directs proteasome-mediated degradation of the tumor suppressor SMAR1 in higher grades of cancer through the anaphase promoting complex. Cell Death Dis 8:e2882. https://doi.org/10.1038/cddis.2017.270
Peng YP et al (2016) The expression and prognostic roles of MCMs in pancreatic cancer. PloS One 11:e0164150. https://doi.org/10.1371/journal.pone.0164150
Press MF et al (1997) HER-2/neu gene amplification characterized by fluorescence in situ hybridization: poor prognosis in node-negative breast carcinomas. J Clin Oncol 15:2894–2904. https://doi.org/10.1200/jco.1997.15.8.2894
Redig AJ, McAllister SS (2013) Breast cancer as a systemic disease: a view of metastasis. J Intern Med 274:113–126. https://doi.org/10.1111/joim.12084
Ren F et al (2015) MYBL2 is an independent prognostic marker that has tumor-promoting functions in colorectal cancer. Am J Cancer Res 5:1542–1552
Rimawi M et al (2018) First-line trastuzumab plus an aromatase inhibitor, with or without pertuzumab, in human epidermal growth factor receptor 2-positive and hormone receptor-positive metastatic or locally advanced breast cancer (PERTAIN): a randomized open-label phase II trial. J Clin Oncol 36:2826–2835. https://doi.org/10.1200/jco.2017.76.7863
Sagara Y et al (2018) Effectiveness of neo-adjuvant systemic therapy with trastuzumab for basal HER2 type breast cancer: results from retrospective cohort study of Japan Breast Cancer Research Group (JBCRG)-C03. Breast Cancer Res Treat 171:675–683. https://doi.org/10.1007/s10549-018-4873-0
Sala A (2005) B-MYB, a transcription factor implicated in regulating cell cycle, apoptosis and cancer. Eur J Cancer (Oxford, England: 1990) 41:2479–2484. https://doi.org/10.1016/j.ejca.2005.08.004
Shen Z et al (2018) CDCA5 regulates proliferation in hepatocellular carcinoma and has potential as a negative prognostic marker. OncoTargets Ther 11:891–901. https://doi.org/10.2147/ott.s154754
Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL (1987) Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science (New York) 235:177–182
Takada M et al (2018) Prediction of postoperative disease-free survival and brain metastasis for HER2-positive breast cancer patients treated with neoadjuvant chemotherapy plus trastuzumab using a machine learning algorithm. Breast Cancer Res Treat. https://doi.org/10.1007/s10549-018-4958-9
Tao D, Pan Y, Jiang G, Lu H, Zheng S, Lin H, Cao F (2015) B-Myb regulates snail expression to promote epithelial-to-mesenchymal transition and invasion of breast cancer cell. Med Oncol (Northwood, London, England) 32:412. https://doi.org/10.1007/s12032-014-0412-y
Thorner AR, Hoadley KA, Parker JS, Winkel S, Millikan RC, Perou CM (2009) In vitro and in vivo analysis of B-Myb in basal-like breast cancer. Oncogene 28:742–751. https://doi.org/10.1038/onc.2008.430
Thu KL et al (2018) Disruption of the anaphase-promoting complex confers resistance to TTK inhibitors in triple-negative breast cancer. Proc Natl Acad Sci USA 115:E1570–e1577. https://doi.org/10.1073/pnas.1719577115
Trainer AH, Thompson E, James PA (2011) BRCA and beyond: a genome-first approach to familial breast cancer risk assessment. Discov Med 12:433–443
Uhlen M et al (2010) Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 28:1248–1250. https://doi.org/10.1038/nbt1210-1248
Uhlen M et al (2015) Proteomics. Tissue-based map of the human proteome. Science (New York) 347:1260419. https://doi.org/10.1126/science.1260419
von Minckwitz G et al (2018) Efficacy and safety of ABP 980 compared with reference trastuzumab in women with HER2-positive early breast cancer (LILAC study): a randomised, double-blind, phase 3 trial. Lancet Oncol 19:987–998. https://doi.org/10.1016/s1470-2045(18)30241-9
Wang J et al (2017) Cdc20 overexpression is involved in temozolomide-resistant glioma cells with epithelial-mesenchymal transition. Cell Cycle (Georget) 16:2355–2365. https://doi.org/10.1080/15384101.2017.1388972
Wang J et al (2018) Silencing of CDCA5 inhibits cancer progression and serves as a prognostic biomarker for hepatocellular carcinoma. Oncol Rep 40:1875–1884. https://doi.org/10.3892/or.2018.6579
Yuan B et al (2006) Increased expression of mitotic checkpoint genes in breast cancer cells with chromosomal instability. Clin Cancer Res 12:405–410. https://doi.org/10.1158/1078-0432.Ccr-05-0903
Zhang H et al (2018a) Acetylation of AGO2 promotes cancer progression by increasing oncogenic miR-19b biogenesis. Oncogene. https://doi.org/10.1038/s41388-018-0530-7
Zhang Z, Shen M, Zhou G (2018b) Upregulation of CDCA5 promotes gastric cancer malignant progression via influencing cyclin E1. Biochem Biophys Res Commun 496:482–489. https://doi.org/10.1016/j.bbrc.2018.01.046
Acknowledgements
The excellent technical assistance of Jianing Tang is gratefully acknowledged.
Funding
This study was supported by National Natural Science Foundation of China (Grant No. 81800429), the Fundamental Research Funds for the Central Universities (Grant No. 2042018kf0065), Health Commission of Hubei Province Scientific Research Project (Grant No. WJ2019Q047), and Zhongnan Hospital of Wuhan University Science, Technology and Innovation Seed Fund (Grant Nos. znpy2017001 and znpy2018028).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, Q., Ren, J., Hou, J. et al. Co-expression network analysis identified candidate biomarkers in association with progression and prognosis of breast cancer. J Cancer Res Clin Oncol 145, 2383–2396 (2019). https://doi.org/10.1007/s00432-019-02974-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-019-02974-4