Abstract
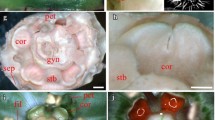
Attractive petals are an integral component of animal-pollinated flowers and in many flowering plant species are restricted to the second floral whorl. Interestingly, multiple times during angiosperm evolution, petaloid characteristics have expanded to adjacent floral whorls or to extra-floral organs. Here, we investigate developmental characteristics of petaloid sepals in Rhodochiton atrosanguineum, a close relative of the model species Antirrhinum majus (snapdragon). We undertook this in two ways, first using scanning electron microscopy we investigate the micromorphology of petals and sepals, followed by expression studies of genes usually responsible for the formation of petaloid structures. From our data, we conclude that R. atrosanguineum petaloid sepals lack micromorphological characteristics of petals and that petaloid sepals did not evolve through regulatory evolution of B-class MADS box genes, which have been shown to specify second whorl petal identity in a number of model flowering plant species including snapdragon. These data, in conjunction with other studies, suggests multiple convergent pathways for the evolution of showy sepals.





Similar content being viewed by others
References
Ambrose B, Lerner D, Ciceri P, Padilla C, Yanofsky M, Schmidt R (2000) Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol Cell 5:569–579
Borchert T, Eckardt K, Fuchs J, Krueger K, Hohe A (2009) 'Who's who' in two different flower types of Calluna vulgaris (Ericaceae): morphological and molecular analyses of flower organ identity. BMC Plant Biol 9:148
Bowman JL (1997) Evolutionary conservation of angiosperm flower development at the molecular and genetic levels. J Biosci 22:515–527
Bowman JL, Smyth DR, Meyerowitz EM (1989) Genes directing flower development in Arabidopsis. Plant Cell 1:37–52
Bowman JL, Smyth DR, Meyerowitz EM (1991) Genetic interactions among floral homeotic genes of Arabidopsis. Development 112:1–20
Brockington SF (2009) Evolution and development of petals in Aizoaceae (Caryophyllales). Dissertation, University of Florida
Brockington SF, Alexandre R, Ramdial J, Moore MJ, Crawley S, Dhingra A, Hilu K, Soltis DE, Soltis PS (2009) Phylogeny of the Caryophyllales sensu lato: revisiting hypotheses on pollination biology and perianth differentiation in the core Caryophyllales. Int J Plant Sci 170:627–643
Brockington SF, Rudall PJ, Frohlich MW, Oppenheimer DG, Soltis PS, Soltis DE (2011) "Living stones" reveal alternative petal identity programs within the core eudicots. Plant J. doi:10.1111/j.1365-313X.2011.04797.x
Broholm SK, Pollanen E, Ruokolainen S, Tahtiharju S, Kotilainen M, Albert VA, Elomaa P, Teeri TH (2010) Functional characterization of B class MADS-box transcription factors in Gerbera hybrida. J Exp Bot 61:75–85
Carpenter R, Coen E (1990) Floral homeotic mutations produced by transposon-mutagenesis in Antirrhinum majus. Genes Dev 4:1483–1493
Christensen K, Hansen H (1998) SEM-studies of epidermal patterns in the angiosperms. Opera Botanica 135:1–91
Coen E, Meyerowitz E (1991) The war of the whorls—genetic interactions controlling flower development. Nature 353:31–37
Coen E, Doyle S, Romero J, Elliott R, Magrath R, Carpenter R (1991) Homeotic genes controlling flower development in Antirrhinum. Development 113:149–155
Cronk Q, Ojeda I (2008) Bird-pollinated flowers in an evolutionary and molecular context. J Exp Bot 59:715–727
De Craene LPR (2007) Are petals sterile stamens or bracts? The origin and evolution of petals in the core eudicots. Ann Bot 100:621–630
De Craene LPR (2008) Homology and evolution of petals in the core eudicots. Syst Bot 33:301–325
de Martino G, Pan I, Emmanuel E, Levy A, Irish VF (2006) Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. Plant Cell 18:1833–1845
Di Stilio VS, Martin C, Schulfer AF, Connelly CF (2009) An ortholog of MIXTA-like2 controls epidermal cell shape in flowers of Thalictrum. New Phytol 183:718–728
Drea S, Hileman LC, De Martino G, Irish VF (2007) Functional analyses of genetic pathways controlling petal specification in poppy. Development 134:4157–4166
Edgar R (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Geuten K, Becker A, Kaufmann K, Caris P, Janssens S, Viaene T, Theissen G, Smets E (2006) Petaloidy and petal identity MADS-box genes in the balsaminoid genera Impatiens and Marcgravia. Plant J 47:501–518
Glover BJ (2007) Understanding flowers and flowering: an integrated approach. Oxford University Press, Oxford
Glover B, Martin C (1998) The role of petal cell shape and pigmentation in pollination success in Antirrhinum majus. Heredity 80:778–784
Goto K, Meyerowitz E (1994) Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev 8:1548–1560
Hileman LC, Irish VF (2009) More is better: the uses of developmental genetic data to reconstruct perianth evolution. Am J Bot 96:83–95
Hileman LC, Sundstrom JF, Litt A, Chen M, Shumba T, Irish VF (2006) Molecular and phylogenetic analyses of the MADS-box gene family in tomato. Mol Biol Evol 23:2245–2258
Howarth D, Baum D (2005) Genealogical evidence of homoploid hybrid speciation in an adaptive radiation of Scaevola (Goodeniaceae) in the Hawaiian Islands. Evolution 59:948–961
Huelsenbeck J, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Irish VF (2009) Evolution of petal identity. J Exp Bot 60:2517–2527
Jack T, Brockman L, Meyerowitz E (1992) The homeotic gene APETALA3 of Arabidopsis thaliana encodes a Mads-box and is expressed in petals and stamens. Cell 68:683–697
Jackson D (1991) In situ hybridization in plants. Mol Plant Pathol Pract Approach 1:163–174
Jaramillo M, Kramer E (2004) APETALA3 and PISTILLATA homologs exhibit novel expression patterns in the unique perianth of Aristolochia (Aristolochiaceae). Evol Dev 6:449–458
Jaramillo MA, Kramer EM (2007) Molecular evolution of the petal and stamen identity genes, APETALA3 and PISTILLATA, after petal loss in the Piperales. Mol Phylogenet Evol 44:598–609
Kang H, Jeon J, Lee S, An G (1998) Identification of class B and class C floral organ identity genes from rice plants. Plant Mol Biol 38:1021–1029
Kanno A, Saeki H, Kameya T, Saedler H, Theissen G (2003) Heterotopic expression of class B floral homeotic genes supports a modified ABC model for tulip (Tulipa gesneriana). Plant Mol Biol 52:831–841
Kay Q, Daoud H, Stirton C (1981) Pigment distribution, light reflection and cell structure in petals. Bot J Linn Soc 83:57–83
Kim S, Yun P, Fukuda T, Ochiai T, Yokoyama J, Kameya T, Kanno A (2007) Expression of a DEFICIENS-like gene correlates with the differentiation between sepal and petal in the orchid, Habenaria radiata (Orchidaceae). Plant Sci 172:319–326
Kramer EM (2007) Understanding the genetic basis of floral diversity. Bioscience 57:479–487
Kramer E, Di Stilio V, Schluter P (2003) Complex patterns of gene duplication in the APETALA3 and PISTILLATA lineages of the Ranunculaceae. Int J Plant Sci 164:1–11
Kramer EM, Holappa L, Gould B, Jaramillo MA, Setnikov D, Santiago PM (2007) Elaboration of B gene function to include the identity of novel floral organs in the lower eudicot Aquilegia. Plant Cell 19:750–766
Krizek B, Meyerowitz E (1996) The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122:11–22
Litt A, Kramer EM (2010) The ABC model and the diversification of floral organ identity. Semin Cell Dev Biol 21:129–137
Liu Y, Nakayama N, Schiff M, Litt A, Irish V, Dinesh-Kumar S (2004) Virus induced gene silencing of a DEFICIENS ortholog in Nicotiana benthamiana. Plant Mol Biol 54:701–711
Maddison D, Maddison W (2005) MacClade 4: analysis of phylogeny and character evolution. 4.08
Maturen N (2008) Genetic analysis of the evolution of petaloid bracts in dogwoods. Dissertation, University of Michigan
Nakamura T, Fukuda T, Nakano M, Hasebe M, Kameya T, Kanno A (2005) The modified ABC model explains the development of the petaloid perianth of Agapanthus praecox ssp. orientalis (Agapanthaceae) flowers. Plant Mol Biol 58:435–445
Noda K, Glover B, Linstead P, Martin C (1994) Flower color intensity depends on specialized cell-shape controlled by a Myb-related transcription factor. Nature 369:661–664
Park J, Ishikawa Y, Yoshida R, Kanno A, Kameya T (2003) Expression of AoDEF, a B-functional MADS-box gene, in stamens and inner tepals of the dioecious species Asparagus officinalis L. Plant Mol Biol 51:867–875
Park J, Ishikawa Y, Ochiai T, Kanno A, Kameya T (2004) Two GLOBOSA-like genes are expressed in second and third whorls of homochlamydeous flowers in Asparagus officinalis L. Plant Cell Physiol 45:325–332
Perez-Rodriguez M, Jaffe F, Butelli E, Glover B, Martin C (2005) Development of three different cell types is associated with the activity of a specific MYB transcription factor in the ventral petal of Antirrhinum majus flowers. Development 132:359–370
Prasad K, Vijayraghavan U (2003) Double-stranded RNA interference of a rice PI/GLO paralog, OsMADS2, uncovers its second-whorl-specific function in floral organ patterning. Genetics 165:2301–2305
Prasad K, Sriram P, Kumar C, Kushalappa K, Vijayraghavan U (2001) Ectopic expression of rice OsMADS1 reveals a role in specifying the lemma and palea, grass floral organs analogous to sepals. Dev Genes Evol 211:281–290
Preston JC, Kellogg EA (2007) Conservation and divergence of APETALA1/FRUITFULL-like gene function in grasses: evidence from gene expression analyses. Plant J 52:69–81
Rasmussen DA, Kramer EM, Zimmer EA (2009) One size fits all? Molecular evidence for a commonly inherited petal identity program in Ranunculales. Am J Bot 96:96–109
Rijpkema AS, Royaert S, Zethof J, van der Weerden G, Gerats T, Vandenbussche M (2006) Analysis of the Petunia TM6 MADS-box gene reveals functional divergence within the DEF/AP3 lineage. Plant Cell 18:1819–1832
Ronquist F, Huelsenbeck J (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Schwarz-Sommer Z, Huijser P, Nacken W, Saedler H, Sommer H (1990) Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 250:931–936
Sharma B, Guo C, Kong H, Kramer EM (2011) Petal-specific subfunctionalization of an APETALA3 paralog in the Ranunculales and its implications for petal evolution. New Phytol 191:870–883
Soltis PS, Brockington SF, Yoo M, Piedrahita A, Latvis M, Moore MJ, Chanderbali AS, Soltis DE (2009) Floral variation and floral genetics in basal Angiosperms. Am J Bot 96:110–128
Sommer H, Beltran JP, Huijser P, Pape H, Lonnig WE, Saedler H, Schwarz-Sommer Z (1990) DEFICIENS, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus—the protein shows homology to transcription factors. EMBO J 9:605–613
Sutton DA (1988) A revision of the tribe Antirrhineae. British Museum Natural History. Oxford University Press, London
Swofford D (2002) PAUP*. Phylogenetic analysis using parsimony (*and other methods) 4
Trobner W, Ramirez L, Motte P, Hue I, Huijser P, Lonnig WE, Saedler H, Sommer H, Schwarz-Sommer Z (1992) GLOBOSA—a homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J 11:4693–4704
Tzeng TY, Yang CH (2001) A MADS-box gene from lily (Lilium longiflorum) is sufficient to generate dominant negative mutation by interacting with PISTILLATA (PI) in Arabidopsis thaliana. Plant Cell Physiol 42:1156–1168
Vandenbussche M, Zethof J, Royaert S, Weterings K, Gerats T (2004) The duplicated B-class heterodimer model: whorl-specific effects and complex genetic interactions in Petunia hybrida flower development. Plant Cell 16:741–754
Vargas P, Rossello JA, Oyama R, Guemes J (2004) Molecular evidence for naturalness of genera in the tribe Antirrhineae (Scrophulariaceae) and three independent evolutionary lineages from the New World and the Old. Plant Syst Evol 249:151–172
Weiss D (2000) Regulation of flower pigmentation and growth: multiple signaling pathways control anthocyanin synthesis in expanding petals. Physiol Plant 110:152–157
Whipple CJ, Ciceri P, Padilla CM, Ambrose BA, Bandong SL, Schmidt RJ (2004) Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development 131:6083–6091
Whitney HM, Glover BJ (2007) Morphology and development of floral features recognised by pollinators. Arthropod-Plant Interact 1:147–158
Whitney HM, Chittka L, Bruce TJA, Glover BJ (2009) Conical epidermal cells allow bees to grip flowers and increase foraging efficiency. Curr Biol 19:948–953
Whittall JB, Voelckel C, Kliebenstein DJ, Hodges SA (2006) Convergence, constraint and the role of gene expression during adaptive radiation: floral anthocyanins in Aquilegia. Mol Ecol 15:4645–4657
Wu H, Su H, Hu J (2007) The identification of A-, B-, C-, and E-class MADS-box genes and implications for perianth evolution in the basal eudicot Trochodendron aralioides (Trochodendraceae). Int J Plant Sci 168:775–799
Xiao H, Wang Y, Liu DF, Wang WM, Li XB, Zhao XF, Xu JC, Zhai WX, Zhu LH (2003) Functional analysis of the rice AP3 homologue OsMADS16 by RNA interference. Plant Mol Biol 52:957–966
Yang YZ, Fanning L, Jack T (2003) The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J 33:47–59
Yoo M, Soltis PS, Soltis DE (2010) Expression of floral MADS-box genes in two divergent water lilies: Nymphaeales and Nelumbo. Int J Plant Sci 171:121–146
Zanis MJ, Soltis PS, Qiu YL, Zimmer E, Soltis DE (2003) Phylogenetic analyses and perianth evolution in basal angiosperms. Ann Mo Bot Gard 90:129–150
Zhang W, Xiang Q, Thomas DT, Wiegmann BM, Frohlich MW, Soltis DE (2008) Molecular evolution of PISTILLATA-like genes in the dogwood genus Cornus (Cornaceae). Mol Phylogenet Evol 47:175–195
Zwickl D (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Dissertation, University of Texas-Austin
Acknowledgments
The authors thank Dr. Jill Preston for the assistance with lab work and discussions on early versions of this manuscript, Dr. David Moore and the imaging facility at the University of Kansas for the help with the SEMs, and Dr. Mark Mort and lab for the help with the phylogenetic analyses. This work was supported by the National Science Foundation (grant IOS-0616025 to L.C.H.).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Schneitz
Rights and permissions
About this article
Cite this article
Landis, J.B., Barnett, L.L. & Hileman, L.C. Evolution of petaloid sepals independent of shifts in B-class MADS box gene expression. Dev Genes Evol 222, 19–28 (2012). https://doi.org/10.1007/s00427-011-0385-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-011-0385-1