Abstract
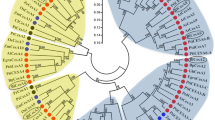
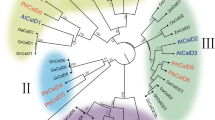
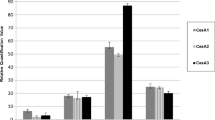
The quality and quantity of cellulose deposited in the primary and secondary cell walls of plants vary in accordance with their biological function. However, the molecular basis of such cellulose heterogeneity has so far remained unclear. Since enrichment of better-quality cellulose, in terms of increased degree of polymerization and crystallinity, is one of the goals of forest biotechnology, our main objective is to decipher the roles of distinct cellulose synthase (CesA) genes in tree development, with special reference to wood production. Here, we report two full-length CesA cDNAs, PtrCesA3 and PtrCesA4, from an economically important tree aspen (Populus tremuloides). PtrCesA3 is orthologous to the Arabidopsis AtCesA4 gene involved in secondary wall formation, whereas PtrCesA4 is orthologous to the Arabidopsis AtCesA1 gene involved in primary cell wall formation. To define the specific cell types expressing these CesA genes, we explored the natural distribution patterns of PtrCesA3 and PtrCesA4 transcripts in a variety of aspen organs, such as leaves, petiole, stem, and roots, using in situ hybridization with hypervariable region-specific antisense riboprobes. Such a side-by-side comparison suggested that PtrCesA3 is exclusively expressed in secondary-wall-forming cells of xylem and phloem fibers, whereas PtrCesA4 is predominantly expressed in primary-wall-forming expanding cells in all aspen organs examined. These findings suggest a functionally distinct role for each of these two types of PtrCesAs during primary and secondary wall biogenesis in aspen trees, and that such functional distinction appears to be conserved between annual herbaceous plants and perennial trees.




Similar content being viewed by others
Abbreviations
- AOR :
-
Acridine orange
- CesA :
-
Cellulose synthase
- DIG :
-
Digoxygenin
- HVR :
-
Hypervariable region
- ORF :
-
Open reading frame
- RT-PCR :
-
Reverse transcription-polymerase chain reaction
- TBO :
-
Toludine blue O
References
Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Hofte H, Plazinski J, Birch R, Cork A, Glover J, Redmond J, Williamson RE (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279:717–720
Carpita N, McCann M (2000) The cell wall. In: Buchanan BB, Gruissem W, Jones RL (eds) Biochemistry and molecular biology of plants. American Society of Plant Physiologists, Rockville, Md., pp 52–108
Delmer DP (1999) Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol 50:245–276
Delmer DP, Amor Y (1995) Cellulose biosynthesis. Plant Cell 7:987–1000
Devereux J, Haeberli P, Smithies O (1984) A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res 12:387–395
Doblin MS, Kurek I, Jacob-Wilk D, Delmer DP (2002) Cellulose biosynthesis in plants: from genes to rosettes. Plant Cell Physiol 43:1407–1420
Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Hofte H (2000) PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12:2409–2423
Haigler C (1985) Functions and biogenesis of native cellulose. In: Nevell TP, Zoronian SH (eds) Cellulose chemistry and applications. Ellis Horwood, Chichester, pp 30–83
Harding SA, Leshkevich J, Chiang VL, Tsai C-J (2002) Differential substrate inhibition couples kinetically distinct 4-coumarate:CoA ligases with spatially distinct metabolic roles in quaking aspen. Plant Physiol 128:428–438
Holland N, Holland D, Helentjaris T, Dhugga K, Xoconostle-Cazares B, Delmer DP (2000) A comparative analysis of the plant cellulose synthase (CesA) gene family. Plant Physiol 123:1313–1323
Joshi CP (2003) Molecular biology of cellulose biosynthesis in plants. In: Pandalai R (ed) Recent research developments plant molecular biology. Research Signpost, Kerala, India, pp 19–38
Kalluri U, Joshi CP (2003) Isolation and characterization of a new, full-Length cellulose synthase cDNA from developing xylem of aspen trees. J Exp Bot 54:2187–2188
Liang X, Joshi CP (2004) Molecular cloning of ten hypervariable regions from cellulose synthase gene superfamily in aspen trees. Tree Physiol 24:543–550
Mellerowicz EJ, Baucher M, Sundberg B, Boerjan W (2001) Unraveling cell wall formation in the woody dicot stem. Plant Mol Biol 47:239–274
O’Brien TP, Feder N, McCully ME (1964) Polychromatic staining of plant cell walls by toluidine blue O. Protoplasma 59:367–373
Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA 93:12637–12642
Richmond T (2000) Higher plant cellulose synthases. Genome Biol 1:reviews3001.1–3001.6
Samuga A, Joshi CP (2002) A new cellulose synthase gene (PtrCesA2) from aspen xylem is orthologous to Arabidopsis AtCesA7 (irx3) gene associated with secondary cell wall synthesis. Gene 296:37–44
Saxena IM, Brown RM, Fevre M, Geremia RA, Henrissat B (1995) Multidomain architecture of β-glycosyltransferases: implications for mechanism of action. J Bacteriol 177:1419–1424
Scheible W-R, Eshed R, Richmond T, Delmer D, Somerville C (2001) Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis ixr1 mutants. Proc Natl Acad Sci USA 98:10079–10084
Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100:1450–1455
Wang H, Loopstra C (1998) Cloning and characterization of a cellulose synthase cDNA (accession no. AF081534) from xylem of hybrid poplar (Populus tremula × Populus alba) (PGR98–179). Plant Physiol 118:1101
Williamson RE, Burn JE, Hocart CH (2002) Towards the mechanism of cellulose synthesis. Trends Plant Sci 7:461–467
Wu L, Joshi CP, Chiang VL (2000) A xylem-specific cellulose synthase gene from aspen (Populus tremuloides) is responsive to mechanical stress. Plant J 22:495–502
Wullschleger SD, Jansson S, Taylor G (2002) Genomics and forest biology: Populus emerges as the perennial favorite. Plant Cell 14:2651–2655
Acknowledgements
We wish to thank Drs. Vincent Chiang, Chung-Jui Tsai and Glenn Mroz for their help and support. We are also grateful to Drs. Takeshi Fujino and Scott Harding for their advice with in situ hybridization procedures and Dr. Laigeng Li for the gift of the xylem cDNA library. This work was supported in part by an NSF-CAREER (IBN-0236492) award, State of Michigan’s Research Excellence Fund and the United States Department of Agriculture’s McIntire Stennis Forestry program.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kalluri, U.C., Joshi, C.P. Differential expression patterns of two cellulose synthase genes are associated with primary and secondary cell wall development in aspen trees. Planta 220, 47–55 (2004). https://doi.org/10.1007/s00425-004-1329-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-004-1329-z