Abstract
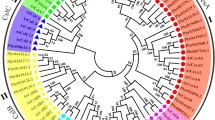
Cellulose, a significant proportion of the plant cell wall, is the main determining factor of the wood qualities of a timber tree. Cellulose synthase (CESA), an essential catalyst for cellulose biosynthesis, is encoded by the CesA gene family. We isolated eight full-length CesA cDNAs from Betula luminifera on the basis of transcriptome sequencing. The predicted proteins comprised 985–1,103 amino acids, sharing 85–92 % identity with the corresponding PtiCESAs from Populus trichocarpa. Each protein contained typical motifs of plant CESA proteins. Phylogenetic analysis showed that BlCESA1, BlCESA3 and BlCESA4 belonged to three clades linked to secondary cell wall synthesis, and BlCESA2, BlCESA5, BlCESA6, BlCESA7 and BlCESA8 belonged to three clades associated with primary cell wall synthesis. Quantitative reverse transcription PCR and in situ mRNA hybridization indicated that transcripts of BlCesA1, BlCesA3 and BlCesA4 were most abundant in the xylem of the lignified stem and had similar expression patterns when treated with hormones and sucrose. Thus, the proteins encoded by these three genes might be subunits of the secondary wall CesA complex. However, the expression patterns of BlCesA1, BlCesA3 and BlCesA4 were different in response to mechanical stress. These three genes showed much higher expression levels in the tension wood than in the control, and they showed different expression patterns in the opposite wood, suggesting that these genes play different roles during secondary wall formation. BlCesA2, BlCesA5, BlCesA6 and BlCesA7 were predominantly expressed in the leaf and phloem. BlCesA8, which is most similar to BlCesA6, was strongly expressed in the leaf, phloem and cambium. Interestingly, these two genes showed distinct expression patterns when treated with hormones, sucrose and bending. Thus, in addition to biosynthesis of the primary cell wall, BlCesA8 might have other functions.






Similar content being viewed by others
References
Appenzeller L, Doblin M, Barreiro R, Wang H, Niu XM, Kollipara K, Carrigan L, Tomes D, Chapman M, Dhugga KS (2004) Cellulose synthesis in maize: isolation and expression analysis of the cellulose synthase (CesA) gene family. Cellulose 11:287–299
Atanassov II, Pittman JK, Turner SR (2009) Elucidating the mechanisms of assembly and subunit interaction of the cellulose synthase complex of Arabidopsis secondary cell walls. J Biol Chem 284:3833–3841
Björklund S, Antti H, Uddestrand I, Moritz T, Sundberg B (2007) Cross-talk between gibberellin and auxin in development of Populus wood: gibberellin stimulates polar auxin transport and has a common transcriptome with auxin. Plant J 52:499–511
Campanella J, Bitincka L, Smalley J (2003) MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinform 4:29
Carraro N, Tisdale-Orr TE, Clouse RM, Knoller AS, Spicer R (2012) Diversification and expression of the PIN, AUX/LAX, and ABCB families of putative auxin transporters in Populus. Front Plant Sci 3:17
Clair B, Ruelle J, Beauchêne J, Prévost MF, Fournier M (2006) Tension wood and opposite wood in 21 tropical rain forest species. 1. Occurence and efficiency of G-layer. Iawa J 27:329–338
Djerbi S, Lindskog M, Arvestad L, Sterky F, Teeri TT (2005) The genome sequence of black cottonwood (Populus trichocarpa) reveals 18 conserved cellulose synthase (CesA) genes. Planta 221:739–746
Endler A, Persson S (2011) Cellulose synthases and synthesis in Arabidopsis. Mol Plant 4:199–211
Engelhardt J (1995) Sources, industrial derivatives, and commercial applications of cellulose. Carbohydr Eur 12:5–14
Festucci-Buselli RA, Otoni WA, Joshi CP (2007) Structure, organization, and functions of cellulose synthase complexes in higher plants. Braz J Plant Physiol 19:1–13
Guerriero G, Fugelstad J, Bulone V (2010) What do we really know about cellulose biosynthesis in higher plants? J Integr Plant Biol 52:161–175
Hamann T, Osborne E, Youngs HL, Misson J, Nussaume L, Somerville C (2004) Global expression analysis of CESA and CSL genes in Arabidopsis. Cellulose 11:279–286
Houben A, Orford SJ, Timmis JN (2006) In situ hybridization to plant tissues and chromosomes. Methods Mol Biol 326:203–218
Huang HH, Xu LL, Tong ZK, Lin EP, Liu QP, Cheng LJ, Zhu MY (2012) De novo characterization of the Chinese fir (Cunninghamia lanceolata) transcriptome and analysis of candidate genes involved in cellulose and lignin biosynthesis. BMC Genom 13:648–661
Joshi CP, Bandari S, Ranjan P, Kalluri UC, Liang X, Fujino T, Samuga A (2004) Genomics of cellulose biosynthesis in poplars. New Phytol 164:53–61
Kalluri UC, Joshi CP (2004) Differential expression patterns of two cellulose synthase genes are associated with primary and secondary cell wall development in aspen trees. Planta 220:47–55
Karpinska B, Karlsson M, Srivastava M, Stenberg A, Schrader J, Sterky F, Bhalerao R, Wingsle G (2004) MYB transcription factors are differentially expressed and regulated during secondary vascular tissue development in hybrid aspen. Plant Mol Biol 56:255–270
Kotake T, Aohara T, Hirano K, Sato A, Kaneko Y, Tsumuraya Y, Takatsuji H, Kawasaki S (2011) Rice brittle culm 6 encodes a dominant-negative form of CesA protein that perturbs cellulose synthesis in secondary cell walls. J Exp Bot 62:2053–2062
Krauskopf E, Harris PJ, Putterill J (2005) The cellulose synthase gene PrCESA10 is involved in cellulose biosynthesis in developing tracheids of the gymnosperm Pinus radiate. Gene 350:107–116
Kumar M, Thammannagowda S, Bulone V, Chiang V, Han KH, Joshi CP, Mansfield SD, Mellerowicz E, Sundberg B, Teeri T, Ellis BE (2009) An update on the nomenclature for the cellulose synthase genes in Populus. Trends Plant Sci 14:248–254
Lin Y, Kao Y-Y, Chen Z-Z, Chu F-H, Chung J-D (2014) cDNA cloning and molecular characterization of five cellulose synthase A genes from Eucalyptus camaldulensis. J Plant Biochem Biotechnol 23(2):199–210
Liu X, Wang Q, Chen P, Song F, Guan M, Jin L, Wang Y, Yang C (2012) Four novel cellulose synthase (CESA) genes from Birch (Betula platyphylla Suk.) involved in primary and secondary cell wall biosynthesis. Int J Mol Sci 13:12195–12212
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(t)) method. Methods 25:402–408
Lu S, Li L, Yi X, Joshi CP, Chiang VL (2008) Differential expression of three eucalyptus secondary cell wall-related cellulose synthase genes in response to tension stress. J Exp Bot 59:681–695
Mutwil M, Debolt S, Persson S (2008) Cellulose synthesis: a complex complex. Curr Opin Plant Biol 11:252–257
Nairn CJ, Haselkorn T (2005) Three loblolly pine CesA genes expressed in developing xylem are orthologous to secondary cell wall CesA genes of angiosperms. New Phytol 166:907–915
Ohashi-Ito K, Demura T, Fukuda H (2002) Promotion of transcript accumulation of novel Zinnia immature xylem-specific HD-Zip III homeobox genes by brassinosteroids. Plant Cell Physiol 43:1146–1153
Palle SR, Candace MS, Seeve CM, Eckert AJ, Cumbie WP, Goldfarb B, Loopstra CA (2011) Natural variation in expression of genes involved in xylem development in loblolly pine (Pinus taeda L.). Tree Genet Genomes 7:193–206
Paux E, Carocha V, Marques C, Mendes de Sousa A, Borralho N, Sivadon P, Grima-Pettenati J (2005) Transcript profiling of Eucalyptus xylem genes during tension wood formation. New Phytol 167:89–100
Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci 93:12082–12085
Persson B, Argos P (1994) Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J Mol Biol 237:182–192
Pfaffl MW (2006) Relative quantification. In: Tevfik Dorak M (ed) Real-time PCR. Taylor & Francis, New York, pp 63–82
Prassinos C, Ko J-H, Yang J, Han K-H (2005) Transcriptome profiling of vertical stem segments provides insights into the genetic regulation of secondary growth in hybrid aspen trees. Plant Cell Physiol 46:1213–1225
Ranik M, Myburg AA (2006) Six new cellulose synthase genes from Eucalyptus are associated with primary and secondary cell wall biosynthesis. Tree Physiol 26:545–556
Richmond T (2000) Higher plant cellulose synthases. Genome Biol 1:1–6
Samuga A, Joshi CP (2002) A new cellulose synthase gene (PtrCesA2) from aspen xylem is orthologous to Arabidopsis AtCesA7 (irx3) gene associated with secondary cell wall synthesis. Gene 296:37–44
Samuga A, Joshi CP (2004) Differential expression patterns of two new primary cell wall-related cellulose synthase cDNAs, PtrCesA6 and PtrCesA7 from aspen trees. Gene 334:73–82
Song D, Shen J, Li L (2010) Characterization of cellulose synthase complexes in Populus xylem differentiation. New Phytol 187:777–790
Suzuki S, Li L, Sun YH, Chiang VL (2006) The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol 142:1233–1245
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4. 0. Mol Biol Evol 24:1596–1599
Tanaka K, Murata K, Yamazaki M, Onosato K, Miyao A, Hirochika H (2003) Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol 133:73–83
Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100:1450–1455
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Timmers J, Vernhettes S, Desprez T, Vincken JP, Visser RG, Trindade LM (2009) Interactions between membrane-bound cellulose synthases involved in the synthesis of the secondary cell wall. FEBS Lett 583:978–982
Ursache R, Nieminen K, Helariutta Y (2013) Genetic and hormonal regulation of cambial development. Physiol Plant 147:36–45
Wang LQ, Guo K, Li Y, Tu YY, Hu HZ, Wang BR, Cui XC, Peng LC (2010) Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol 10:282–297
Wu LG, Joshi CP, Chiang VL (2000) A xylem-specific cellulose synthase gene from aspen (Populus tremuloides) is responsive to mechanical stress. Plant J 22:495–502
Yan C, Yan S, Zeng X, Zhang Z, Gu M (2007) Fine mapping and isolation of Bc7(t), allelic to OsCesA4. J Genet Genomics 34:1019–1027
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China (grant no. 31100494), the Zhejiang Province Science and Technology Support Program (grant nos. 2011C12014 and 2012C12908-8) and Research Center of Bio-Breeding Industry, Zhejiang A & F University (2013SWZY02-01). We appreciate the help given by Youmin Yu of the wood science discipline. We also thank anonymous reviewers for constructive comments on this manuscript.
Conflict of interest
None declared.
Author information
Authors and Affiliations
Corresponding author
Additional information
Huahong Huang, Jiang Cheng and Zaikang Tong have contributed equally to this work. Erpei Lin is the corresponding author.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10570_2014_261_MOESM1_ESM.tif
Tension experiment and sampling of xylem in B. luminifera. A. Tension experiment of B. luminifera. Bent tree with harvesting zone (open box). Tension wood on upper side; opposite wood on lower side. B. Xylem of harvesting zone was divided into three equal parts. The upper was the TW sample; the lower was the OW sample. Scale bars: (A) 5.0 cm, (B) 2.0 cm. (TIFF 5868 kb)
10570_2014_261_MOESM2_ESM.eps
Amino acid alignment and motif analysis of the eight predicted BlCESA protein sequences. Identical conserved residues at a particular site are shown with a black background. A gray background highlights non-identical conserved residues with similar properties. TM, transmembrane region. (EPS 1782 kb)
10570_2014_261_MOESM3_ESM.tif
Sectioning of the up-right current-year branches from B. luminifera. A, B, C and D represent handmade transverse sections of the first, third, fifth and eighth internodes, respectively. Lignified walls stain greenish blue or bright blue, and most primary (non-lignified) walls stain pinkish-purple. co, cortex; ph, phloem; cz, cambial zone; xy, xylem; pi, pith. Scale bars: 40.0 μm. (TIFF 8017 kb)
10570_2014_261_MOESM4_ESM.tif
Relative expression levels of three positive genes in two tissues after treatment with different hormones. (A) Effect of IAA treatments on BlPIN1 expression; (B) effect of GA3 treatments on BlGA20ox1 expression; (C) effect of BR treatments on BlHDZ1 expression. Expression levels in phloem and xylem after 1/2 MS treatment were used individually as a reference and were set at 1. The relative expression level was log2 transformed; > 0 means upregulated, = 0 means unregulated, and < 0 means downregulated. *Gene expression is significantly regulated (p ≤ 0.01, |log2-fold change| ≥ 1). Error bars represent the standard deviation of four biological replicates. (TIFF 5680 kb)
Rights and permissions
About this article
Cite this article
Huang, H., Jiang, C., Tong, Z. et al. Eight distinct cellulose synthase catalytic subunit genes from Betula luminifera are associated with primary and secondary cell wall biosynthesis. Cellulose 21, 2183–2198 (2014). https://doi.org/10.1007/s10570-014-0261-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10570-014-0261-z