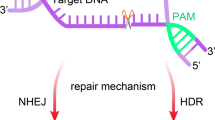
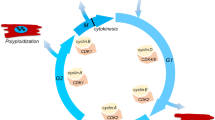
Abstract
Cardiovascular diseases (CVDs) are the leading cause of mortality worldwide. However, the lack of human cardiomyocytes with proper genetic backgrounds limits the study of disease mechanisms. Human pluripotent stem cell–derived cardiomyocytes (hPSC-CMs) have significantly advanced the study of these conditions. Moreover, hPSC-CMs made it easy to study CVDs using genome-editing techniques. This article discusses the applications of these techniques in hPSC for studying CVDs. Recently, several genome-editing systems have been used to modify hPSCs, including zinc finger nucleases, transcription activator-like effector nucleases, and clustered regularly interspaced short palindromic repeat-associated protein 9 (CRISPR/Cas9). We focused on the recent advancement of genome editing in hPSCs, which dramatically improved the efficiency of the cell-based mechanism study and therapy for cardiac diseases.
Graphical abstract






Similar content being viewed by others
Data availability
Not applicable.
References
Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liu DR (2019) Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576:149–157. https://doi.org/10.1038/s41586-019-1711-4
Bultmann S, Morbitzer R, Schmidt CS, Thanisch K, Spada F, Elsaesser J, Lahaye T, Leonhardt H (2012) Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers. Nucleic Acids Res 40:5368–5377. https://doi.org/10.1093/nar/gks199
Chadwick AC, Musunuru K (2017) Genome editing for the study of cardiovascular diseases. Curr Cardiol Rep 19:22. https://doi.org/10.1007/s11886-017-0830-5
Chang L, Zhang J, Tseng YH, Xie CQ, Ilany J, Brüning JC, Sun Z, Zhu X, Cui T, Youker KA, Yang Q, Day SM, Kahn CR, Chen YE (2007) Rad GTPase deficiency leads to cardiac hypertrophy. Circulation 116(25):2976–2983. https://doi.org/10.1161/CIRCULATIONAHA.107.707257
Christidi E, Huang HM, Brunham LR (2018) CRISPR/Cas9-mediated genome editing in human stem cell-derived cardiomyocytes: applications for cardiovascular disease modeling and cardiotoxicity screening. Drug Discov Today Technol 28:13–21. https://doi.org/10.1016/j.ddtec.2018.06.002
Das J, Bhatia P, Singh A (2019) CRISP points on establishing CRISPR-Cas9 in vitro culture experiments in a resource constraint haematology oncology research lab. Indian journal of hematology & blood transfusion: an official journal of Indian Society of Hematology and Blood Transfusion 35(2):208–214. https://doi.org/10.1007/s12288-018-1008-z
Den Hartogh SC, Passier R (2016) Concise review: fluorescent reporters in human pluripotent stem cells: contributions to cardiac differentiation and their applications in cardiac disease and toxicity. Stem Cells 34:13–26. https://doi.org/10.1002/stem.2196
DeWeirdt PC, Sangree AK, Hanna RE et al (2020) Genetic screens in isogenic mammalian cell lines without single cell cloning. Nat Commun 11:752. https://doi.org/10.1038/s41467-020-14620-6
Gaj T, Gersbach CA, Barbas CF (2013) 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31:397–405. https://doi.org/10.1016/j.tibtech.2013.04.004
Gao Y, Wu S, Pan J, Zhang K, Li X, Xu Y, Jin C, He X, Shi J, Ma L, Wu F (2021) CRISPR/Cas9-edited triple-fusion reporter gene imaging of dynamics and function of transplanted human urinary-induced pluripotent stem cell-derived cardiomyocytes. Eur J Nucl Med Mol Imaging 48:708–720. https://doi.org/10.1007/s00259-020-05087-0
Gehrke JM, Cervantes O, Clement MK, Wu Y, Zeng J, Bauer DE, Pinello L, Joung JK (2018) An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat Biotechnol 36:977–982. https://doi.org/10.1038/nbt.4199
Guo R, Wan F, Morimatsu M, Xu Q, Feng T, Yang H, Gong Y, Ma S, Chang Y, Zhang S, Jiang Y (2021) Cell sheet formation enhances the therapeutic effects of human umbilical cord mesenchymal stem cells on myocardial infarction as a bioactive material. Bioactive materials 6:2999–3012. https://doi.org/10.1016/j.bioactmat.2021.01.036
Hernandez MJ, Christman KL (2017) Designing acellular injectable biomaterial therapeutics for treating myocardial infarction and peripheral artery disease. JACC Basic Transl Sci 2:212–226. https://doi.org/10.1016/j.jacbts.2016.11.008
Hinson JT, Chopra A, Nafissi N, Polacheck WJ, Benson CC, Swist S, Gorham J, Yang L, Schafer S, Sheng CC, Haghighi A (2015) Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science 349:982–986. https://doi.org/10.1126/science.aaa5458
Jankele R, Svoboda P (2014) TAL effectors: tools for DNA targeting. Brief Funct Genomics 13:409–419. https://doi.org/10.1093/bfgp/elu013
Jeziorowska D, Korniat A, Salem JE, Fish K, Hulot JS (2015) Generating patient-specific induced pluripotent stem cells-derived cardiomyocytes for the treatment of cardiac diseases. Expert opinion on biological therapy 15(10): pp.1399–1409. https://doi.org/10.1517/14712598.2015.1064109
Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E., Anders C, Hauer M, Zhou K, Lin S, Kaplan M, lavarone AT, Charpentier E, Nogales E, Doudna JA (2014) Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343(6176).
Karakikes I, Termglinchan V, Cepeda DA, Lee J, Diecke S, Hendel A, Itzhaki I, Ameen M, Shrestha R, Wu H, Ma N (2017) A comprehensive TALEN-based knockout library for generating human-induced pluripotent stem cell–based models for cardiovascular diseases. Circ Res 120:1561–1571. https://doi.org/10.1161/CIRCRESAHA.116.309948
Kaur B, Perea-Gil I, Karakikes I (2018) Recent progress in genome editing approaches for inherited cardiovascular diseases. Curr Cardiol Rep 20:58. https://doi.org/10.1007/s11886-018-0998-3
Kim Y, Kweon J, Kim A, Chon JK, Yoo JY, Kim HJ, Kim S, Lee C, Jeong E, Chung E, Kim D, Lee MS, Go EM, Song HJ, Kim H, Cho N, Bang D, Kim S, Kim JS (2013) A library of TAL effector nucleases spanning the human genome. Nat Biotechnol 31:251–258. https://doi.org/10.1038/nbt.2517
Kim YG, Cha J, Chandrasegaran S (1996) Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A 93:1156–1160. https://doi.org/10.1073/pnas.93.3.1156
Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK et al (2007) Cardiomyocytes derived from human embryonic stem cells in prosurvival factors enhance function of infarcted rat hearts. Nat Biotechnol 25(9):1015–1024
Lan F, Lee AS, Liang P, Sanchez-Freire V, Nguyen PK, Wang L, Han L, Yen M, Wang Y, Sun N, Abilez OJ (2013) Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell 12(1):101–113
Lee EJ, Choi EK, Kang SK, Kim GH, Park JY, Kang HJ, Lee SW, Kim KH, Kwon JS, Lee KH, Ahn Y (2012) N-cadherin determines individual variations in the therapeutic efficacy of human umbilical cord blood-derived mesenchymal stem cells in a rat model of myocardial infarction. Mol Ther 20:155–167. https://doi.org/10.1038/mt.2011.202
Lee HK, Willi M, Miller SM, Kim S, Liu C, Liu DR, Hennighausen L (2018) Targeting fidelity of adenine and cytosine base editors in mouse embryos. Nat Commun 9:4804. https://doi.org/10.1038/s41467-018-07322-7
Li X, Lu WJ, Li YN, Wu F, Bai R, Ma S, Dong T, Zhang H, Lee AS, Wang Y, Lan F (2019) MLP-deficient human pluripotent stem cell derived cardiomyocytes develop hypertrophic cardiomyopathy and heart failure phenotypes due to abnormal calcium handling. Cell Death Dis 10:1–15. https://doi.org/10.1038/s41419-019-1826-4)
Li Y.N, Chang Y, Li X, Li X, Gao J, Zhou Y, Wu F, Bai R, Dong T, Ma S, Zhang S (2020) RAD-deficient human cardiomyocytes develop hypertrophic cardiomyopathy phenotypes due to calcium dysregulation. Frontiers in cell and developmental biology 8. https://doi.org/10.3389/fcell.2020.585879.
Li H, Yang Y, Hong W et al (2020) Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Sig Transduct Target Ther 5:1. https://doi.org/10.1038/s41392-019-0089-y
Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP (2013) Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc 8:162–175. https://doi.org/10.1038/nprot.2012.150
Limpitikul WB, Dick IE, Joshi-Mukherjee R, Overgaard MT, George AL Jr, Yue DT (2014) Calmodulin mutations associated with long QT syndrome prevent inactivation of cardiac L-type Ca (2+) currents and promote proarrhythmic behavior in ventricular myocytes. J Mol Cell Cardiol 74:115–124. https://doi.org/10.1016/j.yjmcc.2014.04.022
Lin Q, Zong Y, Xue C, Wang S, Jin S, Zhu Z, Wang Y, Anzalone AV, Raguram A, Doman JL, Liu DR (2020) Prime genome editing in rice and wheat. Nat Biotechnol 38:582–585
Liu GH, Qu J, Suzuki K, Nivet E, Li M, Montserrat N, Yi F, Xu X, Ruiz S, Zhang W, Wagner U, Kim A, Ren B, Li Y, Goebl A, Kim J, Soligalla RD, Dubova I, Thompson J, Yates J 3rd, Esteban CR, Sancho-Martinez I, Izpisua Belmonte JC (2012) Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature 491:603–607. https://doi.org/10.1038/nature11557
Lloyd-Jones D, Adams RJ, Brown TM, Carnethon M, Dai S, De Simone G, Ferguson TB, Ford E, Furie K, Gillespie C (2010) Executive summary: heart disease and stroke statistics-2010 update: a report from the American Heart Association. Circulation 121:948–954. https://doi.org/10.1161/circulationaha.109.192666
Lombardo A, Cesana D, Genovese P et al (2011) Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat Methods 8:861–869. https://doi.org/10.1038/nmeth.1674)
Ma D, Liu F (2015) Genome editing and its applications in model organisms. Genomics Proteomics Bioinformatics 13:336–344. https://doi.org/10.1016/j.gpb.2015.12.001
Matsa E, Ahrens JH, Wu JC (2016) Human induced pluripotent stem cells as a platform for personalized and precision cardiovascular medicine. Physiol Rev 96:1093–1126. https://doi.org/10.1152/physrev.00036.2015
Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY (1997) Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388:882–887. https://doi.org/10.1038/42264
Musunuru K, Sheikh F, Gupta RM, Houser SR, Maher KO, Milan DJ, Terzic A, Wu JC (2018) Induced pluripotent stem cells for cardiovascular disease modeling and precision medicine: a scientific statement from the American Heart Association. Circulation Genomic and Precision Medicine 11(1):43. https://doi.org/10.1161/HCG.0000000000000043
Nguyen Q, Lim KRQ, Yokota T (2020) Genome editing for the understanding and treatment of inherited cardiomyopathies. Int J Mol Sci 21:733. https://doi.org/10.3390/ijms21030733
Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S (2008) Generation of mouse induced pluripotent stem cells without viral vectors. Science 322:949–953. https://doi.org/10.1126/science.1164270
Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ (2008) Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451:141–146. https://doi.org/10.1038/nature06534
Qi T, Wu F, Xie Y, Gao S, Li M, Pu J, Li D, Lan F, Wang Y (2020) Base editing mediated generation of point mutations into human pluripotent stem cells for modeling disease. Frontiers in cell and developmental biology 8:1029. https://doi.org/10.3389/fcell.2020.590581
Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A (2000) Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol 18:399–404. https://doi.org/10.1038/74447
Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK (2012) FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 30:460–465. https://doi.org/10.1038/nbt.2170
Ronaldson-Bouchard K, Ma SP, Yeager K, Chen T, Song L, Sirabella D, Morikawa K, Teles D, Yazawa M, Vunjak-Novakovic G (2018) Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 556(7700):239–243. https://doi.org/10.1038/s41586-018-0016-3
Rosenthal N (1987) Identification of regulatory elements of cloned genes with functional assays. Methods Enzymol 152:704–720. https://doi.org/10.1016/0076-6879(87)52075-4
Shin J, Chen J, Solnica-Krezel L (2014) Efficient homologous recombination-mediated genome engineering in zebrafish using TALE nucleases. Development 141:3807–3818. https://doi.org/10.1242/dev.108019.93
Shinnawi R, Shaheen N, Huber I, Shiti A, Arbel G, Gepstein A, Ballan N, Setter N, Tijsen AJ, Borggrefe M, Gepstein L (2019) Modeling reentry in the short QT syndrome with human-induced pluripotent stem cell–derived cardiac cell sheets. J Am Coll Cardiol 73(18):2310–2324. https://doi.org/10.1016/j.jacc.2019.02.055
Simon CS, Zhang L, Wu T, Cai W, Saiz N, Nowotschin S, Cai CL, Hadjantonakis AK (2018) A Gata4 nuclear GFP transcriptional reporter to study endoderm and cardiac development in the mouse. Biol Open 7(12):036517. https://doi.org/10.1242/bio.036517
Strong A, Musunuru K (2017) Genome editing in cardiovascular diseases. Nat Rev Cardiol 14(1):11. https://doi.org/10.1038/nrcardio.2016.139
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872. https://doi.org/10.1016/j.cell.2007.11.019
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676. https://doi.org/10.1016/j.cell.2006.07.024
Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM (1998) Embryonic stem cell lines derived from human blastocysts. Science 282:1145–1147. https://doi.org/10.1126/science.282.5391.1145
Tohyama S, Fukuda K (2016) Future treatment of heart failure using human iPSC-derived cardiomyocytes. Etiology and Morphogenesis of Congenital Heart Disease pp.25–31. https://doi.org/10.1007/978-4-431-54628-3_4.
Tsien RY (1998) The green fluorescent protein. Annu Rev Biochem 67:509–544. https://doi.org/10.1146/annurev.biochem.67.1.509
Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, Yuan H, Jiang D, Zhang D, Zangi L, Geva J (2014) Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med 20:616–623. https://doi.org/10.1038/nm.3545
Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153:910–918. https://doi.org/10.1016/j.cell.2013.04.025
Wang Y, Zhang WY, Hu S, Lan F, Lee AS, Huber B, Lisowski L, Liang P, Huang M, De Almeida PE, Won JH (2012) Genome editing of human embryonic stem cells and induced pluripotent stem cells with zinc finger nucleases for cellular imaging. Circ Res 111:1494–1503. https://doi.org/10.1161/CIRCRESAHA.112.274969
Wang Z, Su X, Ashraf M, Kim IM, Weintraub NL, Jiang M, Tang Y (2018) Regenerative therapy for cardiomyopathies. J Cardiovasc Transl Res 11:357–365. https://doi.org/10.1007/s12265-018-9807
Wu H, Wang Y, Zhang Y, Yang M, Lv J, Liu J, Zhang Y (2015) TALE nickase-mediated SP110 knockin endows cattle with increased resistance to tuberculosis. Proc Natl Acad Sci U S A 112(13):E1530–E1539. https://doi.org/10.1073/pnas.1421587112
Yong SB, Chung JY, Song Y, Kim YH (2018) Recent challenges and advances in genetically-engineered cell therapy. J Pharm Investig 48:199–208. https://doi.org/10.1007/s40005-017-0381-1
Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318:1917–1920. https://doi.org/10.1126/science.1151526
Zimmermann M, Murina O, Reijns M, Agathanggelou A, Challis R, Tarnauskaitė Ž, Muir M, Fluteau A, Aregger M, McEwan A, Yuan W, Clarke M, Lambros MB, Paneesha S, Moss P, Chandrashekhar M, Angers S, Moffat J, Brunton VG, Hart T, Durocher D (2018) CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions. Nature 559(7713):285–289. https://doi.org/10.1038/s41586-018-0291-z)
Zu Y, Tong X, Wang Z, Liu D, Pan R, Li Z, Hu Y, Luo Z, Huang P, Wu Q, Zhu Z, Zhang B, Lin S (2013) TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat Methods 10:329–331. https://doi.org/10.1038/nmeth.2374
Funding
Funding support from Beijing Natural Science Foundation No. Z190013, National Natural Science Foundation of China Nos. 81970205 and 81,570,215.
Author information
Authors and Affiliations
Contributions
All the authors whose names appear on the submission.
1. Made substantial contributions to the design of the work; appropriately wrote the text and designed images
2. Drafted the work or revised it critically for important intellectual content
3. Approved the version to be published
4. Agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
I give my consent for the publication of identifiable details, which can include images, and details within the text (“Material”) to be published in the above Journal and Article.
Human and animal ethics
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is a special issue on genome editing in health and disease in Pflugers Archi — European Journal of Physiology.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Saleem, A., Abbas, M.K., Wang, Y. et al. hPSC gene editing for cardiac disease therapy. Pflugers Arch - Eur J Physiol 474, 1123–1132 (2022). https://doi.org/10.1007/s00424-022-02751-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00424-022-02751-2