Abstract
The embryonic muscles of the axial skeleton and limbs take their origin from the dermomyotomes of the somites. During embryonic myogenesis, muscle precursors delaminate from the dermomyotome giving rise to the hypaxial and epaxial myotome. Mutant studies for myogenic regulatory factors have shown that the development of the hypaxial myotome differs from the formation of the epaxial myotome and that the development of the hypaxial myotome depends on the latter within the trunk region. The transcriptional networks that regulate the transition of proliferative dermomyotomal cells into the predominantly post-mitotic hypaxial myotome, as well as the eventual patterning of the myotome, are not fully understood. Similar transitions occurring during the development of the neural system have been shown to be controlled by the Atonal family of helix-loop-helix transcription factors. Here, we demonstrate that ATOH8, a member of the Atonal family, is expressed in a subset of embryonic muscle cells in the dermomyotome and myotome. Using the RNAi approach, we show that loss of ATOH8 in the lateral somites at the trunk level results in a blockage of differentiation and thus causes cells to be maintained in a predetermined state. Furthermore, we show that ATOH8 is also expressed in cultured C2C12 mouse myoblasts and becomes dramatically downregulated during their differentiation. We propose that ATOH8 plays a role during the transition of myoblasts from the proliferative phase to the differentiation phase and in the regulation of myogenesis in the hypaxial myotome of the trunk.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Skeletal myogenesis during embryonic development is a highly regulated process. It relies on small populations of specified precursor cells that expand and generate committed cells, which ultimately differentiate into myocytes. A balance between the committed and the differentiated state of the myogenic precursors needs to be achieved. This would allow for the formation of a functional tissue and the maintenance of a sufficient population of precursor cells for subsequent phases of growth or tissue repair. A better understanding of how this balance is maintained and regulated during myogenesis would not only enable us to understand how muscle is formed and maintained during development, but may also be significant in the tailoring of therapeutic approaches for the treatment of muscle diseases.
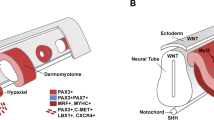
Myotome formation involves an ingression of dermomyotomal myogenic precursors into the myotome from all borders of the dermomyotome. The most recent view on myotome formation proposes that myotomal growth is initiated at the dorsomedial lip of the dermomyotome (DML), which then provides myoblasts that act as a scaffold for later waves of myotomal growth. This theory supports active cellular migration into the myotomal compartment with considerable myocyte contributions from all borders of the somite (Cinnamon et al. 1999; Kahane et al. 2002). Using improved cell lineage and imaging techniques, it was shown that the DML and the ventrolateral lip (VLL) contribute exclusively to the epaxial and hypaxial myotome, respectively. Only incremental growth was shown to occur at the DML and VLL, while myocytes from the rostral and caudal borders contribute to coherent myotomal growth (Gros et al. 2004).
It has been shown that the spatial gene expression of dermomyotomal markers is maintained in the myotome following central dermomyotome dissociation. The cellular identity of the medial and lateral dermomyotome is transferred to the epaxial and hypaxial myotome, respectively (Ahmed et al. 2006). As the VLL contributes exclusively to the hypaxial myotome and the DML solely to the epaxial myotome, it is not surprising that the epaxial myotome growth and patterning is different from that of the hypaxial myotome. Elaborate studies in mutant mice deficient in myogenic regulatory factors have shown that Myf5 is sufficient for the induction and progression of epaxial myogenesis in the absence of MyoD. However, Myf5 is unable to efficiently rescue the delayed and defective hypaxial myogenesis that occurs in the absence of MyoD (Kablar et al. 1997, 1998). Based on the gene expression profile and experimental evidence, the epaxial myotome is not only believed to be more mature than the hypaxial myotome, it is also required for the proper differentiation and patterning of the hypaxial myotome at the trunk level (Kahane et al. 2007). It is speculated that BMP signaling from the lateral plate mesoderm delays myogenic differentiation in the hypaxial myotome (Kahane et al. 2007); however, the intracellular factors that maintain and regulate this delay during hypaxial myotome differentiation are not known.
Our understanding of the mechanism that controls the progression of myogenic precursors to commit to terminal differentiation in both the developmental and regeneration context has greatly improved (reviewed in Bentzinger et al. 2012). On the other hand, the transcriptional networks controlling these fate decisions are still largely unknown. This is not the case in neurogenesis, where the differentiation fate decisions are better described. Work pioneered in Drosophila has led to the identification of a family of transcription factors that control the decision of neural cells to embark on lineage commitment and differentiation. These belong to the Atonal family of helix-loop-helix transcription factors. ATOH8 is a novel member of this family (Jarman et al. 1993; Ledent et al. 2002). In mouse, ATOH8 has been found to contribute to endocrine differentiation by modulating specific aspects of Neurog3 function (Lynn et al. 2008). Detailed examination of ATOH8 shows significant variation in both gene structure and regulatory elements among animals, suggesting a diversification in function (Chen et al. 2011). The murine homolog of ATOH8, Math6, is implicated in the specification and differentiation of cell lineages in the nervous system (Inoue et al. 2001). It has been found that ATOH8 is required for the development of the retina, skeletal muscle, and heart in zebrafish (Yao et al. 2010; Rawnsley et al. 2013). Somitic expression of ATOH8 in the mouse embryo at embryonic stages E9.5 and E12.5 has also been reported (Rawnsley et al. 2013). Our findings further our understanding of the role of ATOH8 in embryonic myogenesis.
We report that the homolog of Atonal, ATOH8, is expressed in both developing embryonic muscle and cultured mouse myoblasts (C2C12). We show that during development, ATOH8 is involved in the transition from a progenitor to the differentiated myogenic fate in the hypaxial myotome of the interlimb region. We propose that ATOH8 is required for the fine-tuning of myogenic differentiation during embryonic hypaxial myotome formation.
Results
Embryonic expression of ATOH8 during avian development
The somitic expression of ATOH8 was first detected at HH10, which thereafter rapidly intensified but maintained its strongest expression in the cranially located somites. From stage HH13 through HH18, ATOH8 transcripts were detected in the lateral regions of the somites (Fig. 1a–c, i). A detailed analysis revealed that the expression domain was confined to the hypaxial myotome and the lateral lip of the dermomyotome (Figs. 1i–k, 2c; Supplementary fig. 1 I, J). ATOH8 transcripts in the hypaxial myotome are expressed prominently at the interlimb level, but faded away at the axial levels where the limb buds are present. Moving from stage HH17 toward HH19, the lateral somitic expression domain spreads medially into the myotome, accompanied by a progressive extension of ATOH8 expression into the more caudally located somites (Fig. 1c, d). The earliest transcripts in the medial myotome and dorsomedial dermomyotomal lip were detected from stage HH19 onward (Fig. 1d, j).
a–h Whole-mount in situ hybridization expression pattern of ATOH8 in chicken embryos from HH stage 13–28. Somitic expression of ATOH8 is shown here at HH13 in the cranial somites (white arrow in a). At HH15, the expression extends caudally (white arrows in a). At later stages (HH17), the somitic expression intensifies caudally and medially (white arrows in c). Note the weak expression of ATOH8 in the limb level hypaxial myotome in comparison with the cervical and interlimb regions of the trunk (white arrows in d–f). At HH 19, ATOH8 is detectable in the dorsomedial lip of cranial somites (white arrowhead in d). At HH21–25, the expression is uniform in the entire myotome. From HH26 to HH28, the medial somitic expression is stronger than in the lateral somites (black arrows in g, h). Expression is also seen in the branchial arches (yellow arrowhead in e), segmental plate (black arrowhead in c), otic placode (black arrowhead in f), and limbs (yellow arrows in f, g). i–n Vibratome sections of in situ hybridized chicken embryos for ATOH8 expression from HH18 to HH26. m, n Double in situ hybridization of ATOH8 (blue)/Pax3 (red) and ATOH8 (blue)/MyoD (red), respectively, at HH23. ATOH8 is expressed in the lateral myotome and the lateral lip of the dermomyotome at HH18 and HH19 (white arrows in i, j). The initiation of the medial somitic expression can be seen at HH19 (white arrowheads in j). At HH20, ATOH8 is expressed over the entire medio-lateral extent of the myotome. Double in situ hybridization with Pax3 shows that ATOH8 is predominantly expressed in the myotome (white arrow in m), while Pax3 is predominantly expressed in the dermomyotome (black arrow in m) and in the intermediate zone of the myotome underlying the central dermomyotome. ATOH8 expression in the myotome (white arrows in n) is overlapping that of MyoD; however, the medially located subectodermal ATOH8 expression domain is MyoD negative (white arrowheads in n)
Silencing ATOH8 in the somites affects the expression of myogenic markers. ATOH8 was silenced using an ATOH8-specific shRNA-EGFP construct targeted to the lateral somite, after which the embryos were reincubated for 24 h. Upon fixation, these embryos were processed for in situ hybridization (ATOH8, Pax3, MyoD, and Myf5). The whole-mount images and section images are of the same embryo. The targeted region is indicated by the EGFP expression in the same embryo (white arrows in a, d, g, j). ATOH8 is specifically silenced in the region of the shRNA treatment (white arrows in b, c). The yellow arrow in c shows the normal expression of ATOH8 at the lateral lip of the dermomyotome on the contralateral side. ATOH8 silencing leads to a downregulation of Myf5 (white arrows in e, f) and MyoD (white arrows in h, i), whereas Pax3 (white arrows in k, l) is upregulated. In contrast, the contralateral side of the same embryo does not show any change in Pax3 expression (m). The dotted lines in b, e, h, and k represent the planes of cross-section shown in c, f, i, and Fig. 3c, respectively. The control EGFP reporter plasmid electroporations did not affect the normal expression pattern of any of the genes tested (Supplementary figure 1 A–H)
Between stages HH21-26, somitic expression of ATOH8 was found throughout the entirety of the myotome (Fig. 1e–g, k, l). At HH26, ATOH8 transcripts were also detected in the subectodermal mesenchyme overlying the neural tube, adjacent to the medial lip of the dermomyotome (Fig. 1g, l). The myotomal expression was still detected at HH28-HH29 (Fig. 1h), but was no longer found by HH30 (data not shown). Double staining showed some overlapping with the MyoD expression domain in the myotome (Fig. 1n), while Pax3 was expressed in the dermomyotome and in the intermediate domain (Ben-Yair and Kalcheim 2005) corresponding to the delaminating central dermomyotome (Fig. 1m).
The effect of ATOH8 silencing on myogenic markers in the somite
To investigate the function of ATOH8 during chicken embryogenesis, we took advantage of the RNAi approach by targeting shRNA plasmids to specific regions of a chicken embryo. We targeted the lateral one-third of the dermomyotome and used EGFP signaling as an indicator of the electroporated region (Figs. 2, 3; Supplementary fig. 1). For our RNAi studies, we analyzed the effects of ATOH8 silencing in the somites at trunk level (Fig. 1e, f). We first determined the inhibitory potential of the ATOH8 shRNA constructs by showing a significant decrease in the expression of ATOH8 corresponding to the EGFP signal (Fig. 2a–c). Next, we monitored the effect of silencing ATOH8 expression. The resulting phenotype was that of a localized decrease in the expression of Myosin Heavy Chain (MHC), a marker of terminally differentiated muscle (Fig. 3a, b). We investigated the mechanism underlying the decrease in terminal muscle differentiation by examining the expression of early myogenic determination markers. Following ATOH8 silencing, we also detected a downregulation of Myf5 and MyoD, correlating with the electroporation site (Fig. 2d–i) and implicating a role for ATOH8 in skeletal myogenesis during embryogenesis. Interestingly, we found a significant upregulation of Pax3 (Figs. 2k–m, 3c) in the targeted area (Fig. 2j). To check whether the Pax3 upregulation is at the site of cATOH8 silencing, we subjected the cross-sections of electroporated and Pax3 stained (in situ hybridization) chicken embryos for immunohistochemistry analysis using anti-GFP antibody. The result confirmed that the Pax3 overexpression is indeed at the site of cATOH8 silencing (Fig. 3f–h). Moreover, at the location where the ATOH8 silencing was performed, we observed a morphologically detectable defect in the formation of myotome from the hypaxial dermomyotome (Fig. 3c, d). On the other hand, normal myotome formation was observed on the untreated side (Fig. 3e), as well as in embryos electroporated with EGFP-only control constructs (Supplementary fig. 1 G, H). Supplementary table 1 provides an overview of the number of embryos analyzed for each gene. Control electroporation using the EGFP-only reporter control plasmid did not alter the normal expression of any of the genes analyzed (Supplementary fig. 1 A–H).
Silencing ATOH8 in the somites affects muscle differentiation and cellular arrangement. The embryos electroporated with ATOH8-specific shRNA-EGFP constructs at the lateral somites were reincubated and processed for immunohistochemistry (myosin heavy chain—MHC). The targeted region is indicated by the EGFP expression (immunohistochemistry) (a). ATOH8 silencing leads to the downregulation of MHC (dotted circle in b) at the targeted site (dotted circle in a). A section through the same embryo presented before in Fig. 2 j–m shows a clear upregulation of Pax3 at the ATOH8 shRNA electroporated region (c). The enlarged view of c shows defective hypaxial myotome formation after ATOH8 knockdown (white dotted line in d). The normal myotome formation is shown on the control side (white dotted line in e). Anti-EGFP immunohistochemistry on the cross-sections of ATOH8 shRNA electroporated and Pax3 stained (in situ hybridization) embryo shows the co-localization of Pax3 upregulation and EGFP expression, which indicates ATOH8 shRNA (white arrows in f, h, black arrow in g). ATOH8 RNAi (visualized here by co-electroporation of constructs in combination with the Tol2-EGFP expression system) in the hypaxial dermomyotome resulted in a remarkable distortion of hypaxial myotome development and caused patterning defects in the myofibers 4 days after electroporation (j). Electroporation of the Tol2 stable expression system alone did not show any distortion of myotome formation (k). Red dashed rectangle (i) indicates the hypaxial region in the embryo shown in j and k. NT neural tube, M myotome
Co-electroporation of ATOH8 shRNA constructs in combination with the Tol2-EGFP expression system (Sato et al. 2007; Kawakami 2007) enabled us to study the long-term effects of ATOH8 silencing in the hypaxial dermomyotome by re-incubating the embryos for four more days. We observed a remarkably distorted patterning of VLL-derived cells normally destined to be myogenic progenitors. This was evidenced by the disordered cellular arrangement (Fig. 3j) compared to the parallel alignment of myofibers in the Tol2-EGFP-electroporated control specimens (Fig. 3k).
These results show that ATOH8 is not only expressed in the hypaxial compartment of the myotome, but also in regions where somite-derived myogenic progenitors are transitioning from a progenitor to a determined myogenic myoblast state. Furthermore, we show that the decreasing ATOH8 levels in somite-derived hypaxial dermomyotome result in defective hypaxial myotome formation and a decrease in the expression of both myogenic markers and MHC. Simultaneously, there was an increase in the expression of dermomyotome-derived progenitor cells.
ATOH8 expression in C2C12 myoblasts
Immunocytological staining was performed on C2C12 myoblasts grown on glass cover slips using ATOH8-specific primary antibodies. Confocal laser scanning microscopy of these cells showed a dotted pattern of endogenous ATOH8 expression in the cytoplasm, as well as in the nucleus (Fig. 4a, b). In order to confirm the presence of the ATOH8 protein, a Western blot was carried out using the cell extracts obtained from proliferating C2C12 myoblasts (Fig. 4c).
ATOH8 expression is regulated during myogenic differentiation
Using a mouse myoblast cell line real-time PCR-based system, the mRNA expression levels of endogenous ATOH8 were quantified against the myogenic markers, Myf5, Myogenin, and Pax7. We exploited our finding that C2C12 cells express ATOH8 and the fact that these cells can be forced to differentiate by changing the culture conditions, which has been well described in the literature (Lawson and Purslow 2000; Clemente et al. 2005).
The mRNA expression levels for each of the genes studied in proliferating C2C12 cells (cultured in normal growth media (T0)) were recorded. These were then considered as reference points to assess any alteration in the expression level of the respective genes during the C2C12 differentiation program.
As expected, we documented an upregulation in the myogenic differentiation genes, Myf5 and Myogenin, following growth in differentiation culture (Fig. 5). In parallel, we observed a transient but significant increase in ATOH8 mRNA on the second day in differentiation conditions, which is consistent with mRNA levels of Myf5 and Myogenin. Following this, as the C2C12 cells continued their differentiation program, the number of ATOH8 transcripts slowly taper off (6 days studied). A similar trend was observed for Myf5 and Myogenin transcripts as well (Fig. 5). Pax7 expression levels, as expected, decreased progressively from T24 (after one day) toward T144 (after 6 days). We also witnessed a transient reduction in ATOH8 transcripts at T24, which could be attributed to the change in the culture media (growth medium to differentiation medium).
qRT-PCR of ATOH8 and three myogenic markers isolated from C2C12 cells induced to differentiate revealed changes in steady state mRNA expression represented as fold ratio measured at 24, 48, 72, and 144 h (T). The fold ratios were compared to the baseline gene expression levels of proliferating C2C12 cells in growth medium (T0). Upon transfer into differentiation medium, a sharp increase in the levels of ATOH8 was seen at 48 h, followed by a progressive decrease in the expression levels at T72 and T144. Myf5 and MyoD expression levels also showed similar dynamics at all compared time points. Pax7 expression levels, as expected, decreased progressively from T24 toward T144. A transient reduction in the number of ATOH8 transcripts at T24 was also observed. Ratios were calculated using the REST software, with highly significant values denoted by asterisks (*p < 0.001)
Thus, the onset of differentiation is accompanied by an immediate increase in ATOH8 expression in C2C12 myoblasts in culture, pointing toward a function of ATOH8 in the transition from the committed to the differentiated state in myogenesis.
Discussion
ATOH8 is a member of the Atonal bHLH transcription factor family. All members of the Atonal superfamily promote the differentiation of specific types of neurons (Yan and Wang 1998; Tomita et al. 2000; Hutcheson and Vetter 2001). In contrast to their well-characterized roles during neurogenesis, the function of the Atonal family is poorly understood in other developmental contexts. In this study, we show that the bHLH transcription factor ATOH8 is prominently expressed in the myotomal compartment during chicken embryogenesis. We demonstrate that reducing the levels of ATOH8 results in a reduction in markers for myogenic determination and terminal differentiation. At the same time, we found an upregulation in the expression of the early premyogenic progenitor marker, Pax3, in the dermomyotome-derived premyogenic cells.
The myotome is the first primitive type of skeletal muscle to form in amniote embryos underneath the dermomyotome (DM). The initiation of myotome formation is attributed to the dorsomedial lip (DML), which is known to lay out the scaffold for the early epaxial myotome (Gros et al. 2004). The myotome is later populated by myoblasts, which enter the myotome in successive waves from all DM borders (Kahane et al. 2002; Hollway and Currie 2005; Yusuf and Brand-Saberi 2012).
The contribution of the DML to hypaxial myotome formation is variable along the craniocaudal axis of the developing embryo. The VLL at limb level disappears (Christ and Ordahl 1995) as a consequence of an epithelio-mesenchymal transition. This releases somitic precursors destined to form limb muscles and vessel endothelia, with minimal contribution to hypaxial myotome formation. The hypaxial myotome is mainly formed by the myogenic precursors from the medial border of the dermomyotome. In contrast, the hypaxial myotome receives no contributions from the medial dermomyotomal lip within the interlimb areas, as has been shown experimentally (Gros et al. 2004; Kahane et al. 2007).
Development of the hypaxial myotome is delayed in the flank region and is only formed after the medial (epaxial) myotome is already established. This delay of myotome maturation in the hypaxial domain is also reflected in the gene expression profile of the hypaxial myotome, in comparison with the epaxial myotome. The medial myotome expresses MyoD, as well as Myf5, in the chicken embryo, whereas the early hypaxial myotome only expresses Myf5 and is devoid of MyoD transcripts. MyoD expression in the hypaxial myotome is only turned on when the medially located myotomal cells extend laterally toward the hypaxial myotome. This indicates that the delay in differentiation of the hypaxial myotome is abrogated by the medial pioneer myoblasts. Experimental manipulation of the expression of MyoD in the epaxial or hypaxial myotome leads to an abnormal myotome formation, fortifying the hypothesis that the medial pioneer myoblasts from the DML are also required for patterning the hypaxial myotome (Kahane et al. 2007). In our expression analysis, we identified some early expression of ATOH8 in the hypaxial myotome from E2.5 (HH13) onwards, corresponding precisely to the hypaxial myotome prior to the expression of MyoD (as reported by Kahane and colleagues). Gradually, while progressing through the early developmental stages, the expression domain of ATOH8 becomes extended medially, and at E3 (HH19), ATOH8 is detectable in the epaxial myotome and DML of cranial somites. This spread of ATOH8 expression occurs progressively along the craniocaudal embryonic axis. At HH13–14, ATOH8 is expressed by the hypaxial myotome cells from the ventrolateral lip (VLL). These then exhibit a delayed differentiation program due to the lack of MyoD transcripts, as compared to their medial counterparts from the DML. It appears that ATOH8 may play a significant role in the differentiation of the hypaxial myotomes, which characteristically delay their differentiation, awaiting patterning cues from the medial myotomal cells. Silencing of ATOH8 in the VLL perturbs the differentiation program of VLL progenitors, leading to the accumulation of myotome-destined muscle precursors at the targeting site. The later spread of ATOH8 into the central and epaxial myotome during normal development is coherent with the entry of myoblasts from the rostral and caudal borders of the DM, as well as from the central DM (Gros et al. 2005). It is therefore tempting to believe that ATOH8 in the myotome may thus be required for the intercalating growth pattern suggested by the model of coherent myotomal growth (Kahane et al. 1998; Cinnamon et al. 1999). The silencing of ATOH8 in zebrafish has been shown to disrupt myoseptum organization, affecting the typical chevron-shaped muscle arrangement (Yao et al. 2010).
The silencing of ATOH8 in the lateral somites within the trunk region results in a decrease in MyoD, Myf5, and MHC, while Pax3 is upregulated. As also suggested by our expression pattern analysis, ATOH8 marks the hypaxial myotomal cells of the trunk. These cells are in a pre-differentiated state before the myoblasts, entering from the DML, have reached their lateral-most positions. The silencing of ATOH8 in the lateral somites probably halts their progression toward differentiation. Additionally, the transfected group of predetermined muscle precursors failed to enter the myotome, remaining Pax3-positive. Moreover, interfering with the expression of ATOH8 in the hypaxial myotome leads to distortion of the hypaxial cell arrangement. Parallel studies performed by our group where we overexpressed ATOH8 in the ventrolateral lip of the dermomyotome, resulted in the notable upregulation of MyoD, accompanied with a distortion/forking of the hypaxial myotome (data not shown). We therefore believe that ATOH8 expression is a necessary intermediate step between the undetermined progenitor status in the dermomyotome and the determined MyoD positive myoblast in the hypaxial myotome.
The data obtained in chicken embryos is in line with our in vitro analyses performed with C2C12 myoblasts, which are derived from murine adult skeletal muscle stem cells. The immunocytochemistry and Western blot analysis show that C2C12 cells also contain ATOH8 protein. Our C2C12 differentiation study followed by real-time PCR to analyze the expression levels of ATOH8 showed that the ATOH8 mRNA is expressed dynamically during the process of differentiation. Our results show that the expression of ATOH8 was transiently upregulated after 48 h in differentiation medium, but significantly downregulated at all subsequently analyzed time points. The expression dynamics of ATOH8 during this process also correlates with the same characteristic myogenic markers for adult myogenesis (Myf5 and MyoG). As further proof of C2C12 myoblast differentiation, the expression level of Pax7, which is a skeletal muscle stem cell marker, became reduced concomitant with the progress of the differentiation program. Murine ATOH8 (MATH6) expression in the hypaxial mouse myotome of embryonic stages 9.5 and 12.5 dpc has recently been reported (Rawnsley et al. 2013). Taken together, the data indicate that ATOH8 has a conserved, temporally restricted role in both embryonic and adult myogenesis, which may also span some species barriers.
Finally, our work shows that ATOH8 is required for the progression of development toward a differentiated state, in keeping with the roles played by other Atonal family members in their respective models. We thus propose that the expression of ATOH8 is controlled by mechanisms that permit the development of a precursor population. The early detection of ATOH8 transcripts in the hypaxial myotome of the trunk highlights yet another differing gene expression profile to add to the known list of markers in the myotome (Cheng et al. 2004; Ahmed et al. 2006). Furthermore, this study contributes to our current understanding of hypaxial muscle formation and may point toward specialized developmental mechanisms governing the formation of the hypaxial myotome as opposed to the epaxial myotome. Additional studies will be required to analyze the regulation of ATOH8 in the early myotome, more specifically, in the context of local signals.
Materials and methods
Chicken embryos and electroporation
Fertilized chicken eggs obtained from a local breeder were incubated at 37 °C and 80 % relative humidity for the required time to obtain stages 17–18HH. The embryo stages were determined according to Hamburger and Hamilton (Hamburger and Hamilton 1992). Embryos from HH10 to HH28 were killed and fixed in 4 % PFA for in situ hybridization. 2.5 ml albumin was removed using a syringe to lower the blastoderm, making the embryo accessible for the electroporation procedure. The upper side of each egg was windowed to visualize the embryos, followed by the partial removal of the extra embryonic membranes. The ATOH8 shRNA constructs (2–3 μg/μl) were mixed with Fast Green solution (Sigma) at a ratio of 2:1 to ease the detection of injection site. For the co-electroporation of ATOH8 shRNA constructs in combination with the Tol2-EGFP expression system, ATOH8 shRNA constructs were mixed with constructs of Tol2-flanked CAGGS-EGFP (pT2 K-CAGGS-EGFP) and CAGGS-transposase (pCAGGS-T2TP) (Sato et al. 2007; Kawakami 2007). The constructs used in the Tol2 stable expression system were received from the laboratory of Koichi Kawakami. Tol2-EGFP-electroporated embryos were reincubated 4 days after electroporation. The DNA constructs were microinjected into the target somites and electroporated as previously described (Scaal et al. 2004; Dai et al. 2005). The electrodes were placed on each side of the microinjected embryo, and five square pulses of 30–55 V, 20 ms width were applied to each embryo. Upon passing current, the plasmid DNA with its negative charge was forced into the cells of tissues adjacent to the anode side. After 24-h reincubation, the EGFP expression in the transgenic embryos was visualized under fluorescence microscopy and photographed. The embryos were further sectioned using a Leica vibratome at a thickness of 35–60 μm. For permanent slides, sections were embedded in Aquatex from Merck.
ATOH8 shRNA constructs
We chose the plasmid-based RNAi strategy to downregulate ATOH8 expression in chicken embryos. Four target sites were selected to prepare the shRNA constructs as described (Dai et al. 2005). Among them, three constructs containing target sequences, GTTGTCCAAACTGGCCATC, GAATAGCCTGTAACTATAT, and GAAGAGCTTCCAGCCAGTTA, were found to be effective, and a cocktail of both constructs was used for subsequent experiments.
Whole-mount in situ hybridization
The EGFP expressing embryos were fixed overnight in 4 % paraformaldehyde at 4 °C. Separate mRNA riboprobes were used to detect the gene expression of ATOH8, MyoD, Myf5, and Pax3. Riboprobes were labeled with a digoxigenin RNA labeling kit from Boehringer, Mannheim, Germany. Whole-mount in situ hybridization was performed as described previously (Nieto et al. 1996) for normal untreated chicken embryos and for electroporated embryos. Double in situ hybridization was performed as previously described (Caprioli et al. 2002).
Immunocytochemistry/immunohistochemistry
Sections of chicken embryo were fixed in 4 % paraformaldehyde at 4 °C overnight and were permeabilized with 0.5 % (v/v) Triton X-100 in PBS. C2C12 cells were grown on glass cover slips and fixed in the same way. Nonspecific antibody binding was blocked using 20 % (v/v) goat serum in PBS. Primary antibodies were applied overnight at 4 °C. Anti-ATOH8 (Sigma-Aldrich, 1:100), anti-MHC (DSHB, 1:200), and anti-GFP (Acris, 1:200) primary antibodies were used. Primary antibodies were visualized with fluorochrome-conjugated secondary antibodies (Molecular Probes) before mounting in DakoCytomation Faramount fluorescent mounting medium containing 100 ng/ml 4,6-diamidino-2-phenylindole (DAPI).
Western blot
Cell lysate from proliferating C2C12 cells was boiled in 1 × Laemmli buffer for 5 min, separated by 10 % SDS–PAGE gel and blotted onto the nitrocellulose membrane. Blotted membranes were incubated with anti-ATOH8 primary antibody overnight. The blots were then incubated with HRP-conjugated secondary antibody for 1–2 h and the signal detected by SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Microscopy and imaging
After in situ hybridization, samples were observed and photographed using the Leica MZFLIII microscope and Leica DC 300F digital camera. Hybridized sections were observed and photographed using an Axioscope 20 (Zeiss) and a Leica DFC320 digital camera. Images of C2C12 myoblasts were captured using confocal laser scanning microscopy (Zeiss LSM 510 META) in the multi-tracking sequential mode set for the detection of AF-488 and DAPI with the appropriate emission bandpass filters.
RNA preparation, reverse transcription, and quantitative real-time PCR of C2C12 cells
Total RNA was isolated from C2C12 cells using standard TRIzol method (Invitrogen). For real-time PCR, the first strand cDNA was synthesized using PrimeScript Reverse Transcriptase (TaKaRa, Japan). To confirm the successful cDNA synthesis, PCR was performed using primers designed against 18S RNA. SYBR Advantage qPCR mix (Clontech) was used to perform the real-time PCR. The primers were designed against mouse ATOH8, Myogenin, Myf5, and Pax7 genes (sense mATOH8 5′-TCAGCTTCTCCGAGTGTGTG-3 antisense mATOH8 5′-TAGCCTGTGGCAGGTCACCT-3 sense mMyogenin 5′-GAAGCGCAGGCTCAGAAAGT-3′ antisense mMyogenin 5′-GATTGTGGGCGCTGTAGGGT-3′, sense mMyf5 5′-GACAGGGCTGTTACATTCAGG-3′ antisense mMyf5 5′-TGAGGGAACAGGTGGAGAAC-3′, sense mPax7 5′-GTCGGGTTCTGATTCCACAT-3′ antisense mPax7 5′- GCGAGAAGAAAGCCAAACAC-3′). Amplification reactions were performed in triplicates with the primer pairs producing single amplification products with a calculated Tms > 80 °C, which was verified by melting point analysis. Syber Green I reaction conditions were as recommended by the manufacturer (Clontech Mountain View, CA, USA) using the DNA Engine Opticon 2 Real-Time PCR Detection System (Bio-Rad Laboratories GmbH, München, Germany). The threshold cycle (C t) was defined at the point where the fluorescence signal reached a value of 0.01 above background during the exponential phase of the reaction. The average C t values were used to calculate the ratios for all the genes studied using the equation procedure introduced by Pfaffl (Pfaffl et al. 2002). Ratios (R) represented the relative expression of a target gene in a sample:
Primer efficiencies (E) of the PCRs were determined directly from amplification curves using the DNA Engine Opticon 2 Real-Time PCR Detection System software. The C t values for the reference gene 18s rRNA were used to normalize mRNA levels of the samples. The upstream primer (5′-CATGGTGACCACGGGTGAC-3′) and the downstream primer (5′-TTCCTTGGATGTGGTAGCCG-3′) for 18s rRNA were previously reported (Ray et al. 2005). The changes in the mRNA expression levels were calculated as ratios relative to the mRNA levels found at time point T0 of C2C12 differentiation. All experiments represented three independent sets of samples analyzed in triplicate. Statistical analysis was performed using the Relative Expression Software Tool (REST) software (Pfaffl et al. 2002) and the data expressed as expression ratios following the Pair Wise Fixed Reallocation Randomisation Test (Bustin et al. 2005).
References
Ahmed MU, Cheng L, Dietrich S (2006) Establishment of the epaxial-hypaxial boundary in the avian myotome. Dev Dyn 235:1884–1894
Bentzinger CF, Wang YX, Rudnicki MA (2012) Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol 4:a008342
Ben-Yair R, Kalcheim C (2005) Lineage analysis of the avian dermomyotome sheet reveals the existence of single cells with both dermal and muscle progenitor fates. Development 132:689–701
Bustin SA, Benes V, Nolan T, Pfaffl MW (2005) Quantitative real-time RT-PCR—a perspective. J Mol Endocrinol 34:597–601
Caprioli A, Goitsuka R, Pouget C, Dunon D, Jaffredo T (2002) Expression of Notch genes and their ligands during gastrulation in the chicken embryo. Mech Dev 116:161–164
Chen J, Dai F, Balakrishnan-Renuka A, Leese F, Schempp W, Schaller F, Hoffmann MM, Morosan-Puopolo G, Yusuf F, Bisschoff IJ et al (2011) Diversification and molecular evolution of ATOH8, a gene encoding a bHLH transcription factor. PLoS ONE 6:e23005
Cheng L, Alvares LE, Ahmed MU, El-Hanfy AS, Dietrich S (2004) The epaxial–hypaxial subdivision of the avian somite. Dev Biol 274:348–369
Christ B, Ordahl CP (1995) Early stages of chick somite development. Anat Embryol 191:381–396
Cinnamon Y, Kahane N, Kalcheim C (1999) Characterization of the early development of specific hypaxial muscles from the ventrolateral myotome. Development 126:4305–4315
Clemente CF, Corat MA, Saad ST, Franchini KG (2005) Differentiation of C2C12 myoblasts is critically regulated by FAK signaling. Am J Physiol Regul Integr Comp Physiol 289:R862–R870
Dai F, Yusuf F, Farjah GH, Brand-Saberi B (2005) RNAi-induced targeted silencing of developmental control genes during chicken embryogenesis. Dev Biol 285:80–90
Gros J, Scaal M, Marcelle C (2004) A two-step mechanism for myotome formation in chick. Dev Cell 6:875–882
Gros J, Manceau M, Thome V, Marcelle C (2005) A common somitic origin for embryonic muscle progenitors and satellite cells. Nature 435:954–958
Hamburger V, Hamilton HL (1992) A series of normal stages in the development of the chick embryo. 1951. Dev Dyn 195:231–272
Hollway G, Currie P (2005) Vertebrate myotome development. Birth Defects Res C Embryo Today 75:172–179
Hutcheson DA, Vetter ML (2001) The bHLH factors Xath5 and XNeuroD can upregulate the expression of XBrn3d, a POU-homeodomain transcription factor. Dev Biol 232:327–338
Inoue C, Bae SK, Takatsuka K, Inoue T, Bessho Y, Kageyama R (2001) Math6, a bHLH gene expressed in the developing nervous system, regulates neuronal versus glial differentiation. Genes Cells 6:977–986
Jarman AP, Grau Y, Jan LY, Jan YN (1993) Atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell 73:1307–1321
Kablar B, Krastel K, Ying C, Asakura A, Tapscott SJ, Rudnicki MA (1997) MyoD and Myf-5 differentially regulate the development of limb versus trunk skeletal muscle. Development 124:4729–4738
Kablar B, Asakura A, Krastel K, Ying C, May LL, Goldhamer DJ, Rudnicki MA (1998) MyoD and Myf-5 define the specification of musculature of distinct embryonic origin. Biochem Cell Biol 76:1079–1091
Kahane N, Cinnamon Y, Kalcheim C (1998) The origin and fate of pioneer myotomal cells in the avian embryo. Mech Dev 74:59–73
Kahane N, Cinnamon Y, Kalcheim C (2002) The roles of cell migration and myofiber intercalation in patterning formation of the postmitotic myotome. Development 129:2675–2687
Kahane N, Ben-Yair R, Kalcheim C (2007) Medial pioneer fibers pattern the morphogenesis of early myoblasts derived from the lateral somite. Dev Biol 305:439–450
Kawakami K (2007) Tol2: a versatile gene transfer vector in vertebrates. Genome Biol 8(Suppl 1):S7
Lawson MA, Purslow PP (2000) Differentiation of myoblasts in serum-free media: effects of modified media are cell line-specific. Cells Tissues Organs 167:130–137
Ledent V, Paquet O, Vervoort M (2002) Phylogenetic analysis of the human basic helix-loop-helix proteins. Genome Biol 3, RESEARCH0030
Lynn FC, Sanchez L, Gomis R, German MS, Gasa R (2008) Identification of the bHLH factor Math6 as a novel component of the embryonic pancreas transcriptional network. PLoS ONE 3:e2430
Nieto MA, Patel K, Wilkinson DG (1996) In situ hybridization analysis of chick embryos in whole mount and tissue sections. Methods Cell Biol 51:219–235
Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36
Rawnsley DR, Xiao J, Lee J, Liu X, Mericko-Ishizuka P, Kumar V, He J, Basu A, Lu M, Lynn FC et al (2013) The transcription factor Atonal homolog 8 regulates Gata4 and Friend of Gata-2 during vertebrate development. J Biol Chem 288(34):24429–24440
Ray A, Zoidl G, Weickert S, Wahle P, Dermietzel R (2005) Site-specific and developmental expression of pannexin1 in the mouse nervous system. Eur J Neurosci 21:3277–3290
Sato Y, Kasai T, Nakagawa S, Tanabe K, Watanabe T, Kawakami K, Takahashi Y (2007) Stable integration and conditional expression of electroporated transgenes in chicken embryos. Dev Biol 305:616–624
Scaal M, Gros J, Lesbros C, Marcelle C (2004) In ovo electroporation of avian somites. Dev Dyn 229:643–650
Tomita K, Moriyoshi K, Nakanishi S, Guillemot F, Kageyama R (2000) Mammalian achaete–scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system. EMBO J 19:5460–5472
Yan RT, Wang SZ (1998) Identification and characterization of tenp, a gene transiently expressed before overt cell differentiation during neurogenesis. J Neurobiol 34:319–328
Yao J, Zhou J, Liu Q, Lu D, Wang L, Qiao X, Jia W (2010) Atoh8, a bHLH transcription factor, is required for the development of retina and skeletal muscle in zebrafish. PLoS ONE 5:e10945
Yusuf F, Brand-Saberi B (2012) Myogenesis and muscle regeneration. Histochem Cell Biol 138:187–199
Acknowledgments
We thank Prof. Koichi Kawakami, National institute of genetics, Japan for sharing the Tol2-EGFP expression system. We thank Ellen Gimbel, Susanna Glaser, and Ulrike Pein at the Institute of Anatomy and Cell Biology, University of Freiburg, and Eva-Maria Konieczny, Rana Houmany, and Swantje Wulf at the Institute of Anatomy, Ruhr University Bochum for their excellent technical assistance. We also acknowledge the support of “Molecular mechanisms of migration, invasion and metastasis,” a Baden-Württemberg grant, the MYORES Project (511978) funded by the EU’s sixth framework program, GRK1104 of Faculty of Biology, University of Freiburg and, FoRUM F647-09 and F732N-2011 of Faculty of Medicine, Ruhr University Bochum. We thank Daniel Terheyden-Keighley for his help during the preparation of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
A. Balakrishnan-Renuka and G. Morosan-Puopolo have contributed equally to the manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.

418_2013_1155_MOESM1_ESM.jpg
Supplementary Figure 1. The control EGFP reporter plasmid electroporations did not affect the normal expression pattern of any of the genes tested (A-H). During normal development (HH17 shown here), ATOH8 expression is observable at the lateral lip of the dermomyotome (I). The enlarged view of ATOH8 expression (marked by dashed rectangle in I) at the lateral dermomyotomal lip is shown in J (black arrows in J). (JPEG 1128 kb)

418_2013_1155_MOESM2_ESM.jpg
Supplementary Table 1. States the number of embryos electroporated with the ATOH8-specific shRNA-EGFP construct and analyzed using in situ hybridization for the expression of ATOH8, Myf5, MyoD and Pax3. The percentages of observed effects are included. (JPEG 39 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Balakrishnan-Renuka, A., Morosan-Puopolo, G., Yusuf, F. et al. ATOH8, a regulator of skeletal myogenesis in the hypaxial myotome of the trunk. Histochem Cell Biol 141, 289–300 (2014). https://doi.org/10.1007/s00418-013-1155-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-013-1155-0