Abstract
The endoplasmic reticulum (ER) is a key organelle of the secretion pathway involved in the synthesis of both proteins and lipids destined for multiple sites within and without the cell. The ER functions to both co- and post-translationally modify newly synthesized proteins and lipids and sort them for housekeeping within the ER and for transport to their sites of function away from the ER. In addition, the ER is involved in the metabolism and degradation of specific xenobiotics and endogenous biosynthetic products. A variety of proteomics studies have been reported on different subcompartments of the ER providing an ER protein dictionary with new data being made available on many protein complexes of relevance to the biology of the ER including the ribosome, the translocon, coatomer proteins, cytoskeletal proteins, folding proteins, the antigen-processing machinery, signaling proteins and proteins involved in membrane traffic. This review examines proteomics and cytological data in support of the presence of specific molecular machines at specific sites or subcompartments of the ER.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
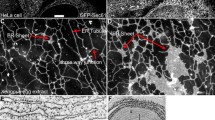
The ER is a dynamic organelle essential for cell life. The main subdomains of the ER include the nuclear envelope (NE), rough ER (rER), and transitional ER (tER), each with its own characteristic structure and function. The NE forms a shell around the nucleus. The shell typically consists of a flattened saccule closely applied to nuclear chromatin and showing multiple discontinuities or nuclear pores (Fig. 1a). The rER is continuous with the NE and consists mostly of stacked, flattened saccules. Each saccule is limited by membranes with attached ribosomal particles (Fig. 1b). The tER is composed of two distinct but continuous membrane domains: a rough domain characterized by the presence of attached ribosomes and a smooth domain (sER), giving rise to membrane buds and tubules (Fig. 1c). Clusters of vesicles, and tubules are often observed closely associated with the tER and represent the intermediate compartment, also called ER-Golgi Intermediate Compartment (ERGIC), vesiculo-tubular clusters (VTCs) or pre-Golgi intermediates (Bannykh et al. 1996; Hauri et al. 2000; Saraste and Kuismanen 1992; Fan et al. 2003). In certain cell types (such as steroid-synthesizing cells or hepatocytes) the smooth ER is extensive and consists of a network of interconnecting tubules limited by smooth membranes devoid of attached ribosomes. The tubules are linked by tripartite junctions and limit cytoplasmic regions often devoid of large organelles (Fig. 1d).
Different subcompartments of the endoplamic reticulum. The ER is composed of continuous but distinct subdomains. a The nuclear envelope (NE) is shown with nuclear pores and ribosomal particles attached to the outer membrane. b The rough ER (rER) is continuous with the NE and consists of stacked flattened saccules, whose limiting membranes have numerous attached ribosomal particles. c Transitional ER (tER) is composed of a rER subdomain continuous with the rER and a smooth ER (sER) subdomain consisting of buds and tubules devoid of associated ribosomes (arrowhead points to a coated bud). d In some cells (e.g., steroid secreting cells and hepatocytes) the sER is composed of a large network of interconnecting tubules showing tripartite junctions (arrows) and fenestrations. Micrograph in C is courtesy of Christian Zuber and Jurgen Roth
The diversity of ER structures parallels its many functions. The numerous ribosomes associated with the flattened saccules of the rER indicate its high capacity for protein synthesis, translocation, and folding. The two domains of the tER indicate an additional function within this compartment: that of the formation of tubules and vesicles allowing cargo exit and transport to the Golgi Apparatus, and the tubules of the sER, which are continuous with the rER indicate an increased ER volume thus providing enhanced capacity for calcium storage, drug handling, detoxification, and lipid and steroid synthesis (reviewed in Baumann and Walz 2001; Shibata et al. 2006). Based on these considerations, the subcompartments of the ER are predicted to have a presence and enrichment of very specific proteins. Such predictions can be analyzed by proteomics analysis of each ER subcompartment.
A number of proteomics studies were done on the subcompartments of the ER providing an ER protein dictionary with new data being made available on many molecular protein complexes of relevance to the biology of the ER. As summarized in Table 1, proteomics studies were done on the NE, total ER microsomes (mixture of rough and smooth ER membranes), ribosomes, purified rough microsomes, purified smooth microsomes, and the ERGIC compartment. Because of the paucity of proteomics data on pure outer nuclear envelope membrane, we have restricted comment on studies of this ER compartment to that within Table 1. Subcompartments of the ER were isolated by subcellular fractionation using centrifugation and various sucrose gradients and some were isolated by immuno-isolation. The degree of purity of these fractions was analyzed by morphology and/or morphometry and in many cases biochemically by measuring the level of enrichment of expected proteins (markers) compared to the starting material (Table 1). Identifying the proteins present in these ER subfractions is an important step towards better understanding the specialized functions of the ER subcompartments and molecular mechanisms governing these functions.
Protein synthesis and secretion
The ER is a key organelle of the secretion pathway involved in the synthesis of both proteins and lipids destined for multiple sites within and without the cell.
Ribosomal proteins
Because ribosomes define the rough domain of the ER, studies of the proteins of purified ribosomes are relevant to the proteome of the rough ER. Link et al. (1999) reported on the proteome of ribosomes purified from Saccharomyces cerevisiae. Multidimensional chromatography and tandem MS were used to identify 70 of the 78 predicted ribosomal proteins in yeast. The YMR116p protein (homologue of human RACK1 an intracellular receptor for activated protein kinase C) was found to be associated with the 40S ribosomal subunit implicating this protein in translation in PKC-mediated signal transduction. RACK1 is now known to be a bona fide ribosomal protein and to play an important role in regulating eukaryotic translation (Nilsson et al. 2004). Of the proteomics studies carried out so far on mammalian liver ER subcompartments, Gilchrist et al. (2006) have identified most of the ribosomal proteins in purified rough microsomes from rat liver including 33 proteins of the 40S ribosomal subunit and 45 proteins of the 60S ribosomal subunit. This number is close to the 84 proteins isolated from rat ribosomal particles (Wool et al. 1995). The study by Gilchrist et al. (2006) revealed that the concentration of ribosomal proteins was greatest in high-density rough microsomes (HDM) when compared with the ribosomal protein concentration in a smooth microsomal fraction containing low-density rough microsomes (LDM). This is consistent with previous morphometric studies showing more ribosomal particles associated with more vesicles in the HDM fraction compared to that in the LDM fraction from normal rat liver (Gilchrist et al. 2006; Lavoie et al. 1996). Foster et al. (2006) identified a comparable number of ribosomal proteins in ER fractions enriched with the marker proteins calnexin and p115.
Proteins involved in RNA metabolism
Rough microsomal derivatives of the ER are expected to contain a variety of proteins involved in RNA metabolism because they contain and translate associated messenger RNA (Lerner and Nicchitta 2006). A variety of proteins known to be involved in the metabolism of mRNA and in translation of mRNA have been detected in ER membrane derivatives by mass spectrometry (Foster et al. 2006; Gilchrist et al. 2006). These include heterogeneous nuclear ribonucleoproteins (hnRNPs, hnRNP A1, hnRNP A2/B1, hnRNP D, and hnRNP K), Poly(A)-binding protein 1 and many proteins that are involved in translation (eEF-1A, eEF-2, several subunits of eIF-3, and eIF-5A). Heterogeneous nuclear ribonucleoproteins are involved in mRNA metabolism both inside the nucleus and in the cytoplasm. In the cytoplasm hnRNPs are known to regulate mRNA localization, mRNA translation, and mRNA turnover (Dreyfuss et al. 2002; Shyu and Wilkinson 2000). Since hnRNP family proteins are known to accompany RNA from the gene through nuclear pores and into polysomes (Visa et al. 1996) the hnRNP family proteins detected in ER maybe associated with membrane-bound polysomes and modulate translation of specific proteins relevant to protein synthesis.
Proteins mediating targeting, co-translational translocation, and processing of nascent polypeptide chains
Secretory and transmembrane proteins are synthesized on polysomes that are attached to the rough domain of the ER (Palade 1975). Ribosomes are bound to the ER membrane and are associated with a molecular complex involved in protein translocation (Gilmore 1993; Rapoport et al. 1996). Sec61p made up of the three subunits alpha, beta, and gamma comprises the main protein-conducting channel, whereas signal-recognition particle receptor, signal peptidase, oligosaccharyltransferase, TRAM, TRAP, p180, p34, Sec63, and BIP comprise associated protein complexes that assist in the signal sequence-mediated targeting, co-translational translocation, and processing of nascent polypeptide chains (Johnson and van Waes 1999; Nilsson et al. 2003; Osborne et al. 2005). As would be expected because of the continuity between rough and smooth ER membrane compartments and because of the capacity of translocon components to diffuse laterally along the ER membrane (Nikonov et al. 2002) translocon components were detected in microsomal-derivatives of both rough and smooth ER (Gilchrist et al. 2006). However, despite such membrane continuity polysomes are retained concentrated in the rough domain of the ER. Cytoskeletal elements (i.e., microtubules) known to interact with specific receptors of the ER (i.e., CLIMP-63) might play a role in mechanisms that restrict the lateral mobility of membrane-bound polysomes (Farah et al. 2005; Vedrenne et al. 2005; Nikonov et al. 2007).
Proteins involved in glycosylation and the calnexin cycle
During the process of protein translocation, the asparagine residues in Asn-x-Ser/Thr motifs on the nascent polypeptide chain are glycosylated in the lumen of the ER by a multisubunit membrane protein complex called oligosaccharyltransferase (OST). Although several subunits (mainly the ribophorins I and II) of OST have previously been identified in proteomics studies of ER subfractions the most complete analysis of these was reported by Shibatani et al. (2005) who carried out proteomics analysis of isolated ribosome-associated membrane protein from canine rough microsomes. These investigators identified five known subunits of OST (STT3-A, ribophorin I, ribophorin II, OST48, and DAD1) and two previously uncharacterized proteins that co-purified with these subunits DC2 and KCP2. Oligosaccharyltransferase transfers preassembled glucose3-mannose9-N-acetylglucosamine2 core oligosaccharides from the ER membrane lipid donor dolichol pyrophosphate to nascent polypeptide chains. Glycosylation facilitates protein folding by increasing the solubility of yet-unstructured nascent chains and allows nascent chains to enter the folding sensor system by recruiting the lectin chaperones calnexin and calreticulin (Ellgaard et al. 1999). Access to the calnexin/calreticulin system requires modification of the N-glycans by the enzymes glucosidase I (GI), glucosidase II (GII), UDPglucose: glycoprotein glucosyltransferase (UGT1), ER-mannosidase 1 and EDEMs (ER degradation enhancing-mannosidase-like protein) (reviewed by Ruddock and Molinari 2006). Polypeptide released from this sensor system fulfils quality control requirements and can exit ER and transport to their final destination. However, misfolded polypeptides are retrotranslocated into the cytosol and degraded (described in “Ubiquitin metabolizing enzymes”). A number of enzymes involved in N-glycan modification have been reported in association with ER subfractions including GI (Gilchrist et al. 2006), GII (Knoblach et al. 2003; Foster et al. 2006; Gilchrist et al. 2006) and UGT1 (Gilchrist et al. 2006) following analysis by mass spectrometry. In addition to the expected detection in the rough microsomes (Knoblach et al. 2003; Gilchrist et al. 2006), these enzymes have also been detected in smooth microsomes (Gilchrist et al. 2006). This is consistent with the localization of GII and UGT in smooth ER and also ERGIC by immuno-electron microscopy (reviewed by Roth et al. 2002). These data are also consistent with the detection of chaperones calnexin and calreticulin, in rough and smooth microsomes from rat liver (Gilchrist et al. 2006) as well as the presence the ERAD machinery in smooth microsomes (Gilchrist et al. 2006). Thus, glycoprotein quality control not only takes place in the rER but also may occur to a significant extent in the sER.
Biosynthetic cargo
Many hepatic secretory proteins were detected by proteomics analyses within ER fractions from normal liver. These include albumin, serotransferrin, apolipoproteins A, B, and E, Complement C3, alpha-2u-globulin, and transthyretin (Foster et al. 2006; Galeva and Altermann 2002; Gilchrist et al. 2006; Knoblach et al. 2003). The association of these proteins with the endoplasmic reticulum is assumed to be biogenetic. The proteins are thought to be in the lumen of the ER en route for secretion via the early secretion apparatus. Consistent with this suggestion quantitative proteomics revealed two to fivefold higher concentrations of these proteins in a Golgi fraction when compared with that in rough and smooth ER fractions from the same tissue (Gilchrist et al. 2006). Although transmembrane proteins destined for transport out of the ER to the Golgi Apparatus and to the plasma membrane are expected to be part of the biosynthetic cargo of the ER such proteins maybe either too low in abundance for detection by mass spectrometry or they could be considered as membrane contaminants of the ER preparations. Confirmation of the latter suggestion would require double labeling for specific membrane markers using imunocytochemistry of the subcellular fractions used for proteomics analysis.
Proteins involved in cargo exit and membrane traffic
Interactions between membrane derivatives of the ER and the Golgi Apparatus are well known and this includes formation of ER exit sites to permit transport of newly synthesized protein out of the ER towards the Golgi (Palade 1975) and the formation of the ER-Golgi intermediate compartment to permit membrane recycling between the ER and the Golgi (Lee et al. 2004). Proteomics studies of different fractions of the ER have detected a number of proteins involved in membrane traffic. Coat proteins of the COPI and COPII protein complexes were detected in smooth microsomes (Gilchrist et al. 2006) and in ER/Golgi/vesicles (Foster et al. 2006). The smooth microsomes used in the studies of Gilchrist et al. (2006) are considered derivatives of the tER based on previous molecular, histochemical and morphological characterizations (Lavoie et al. 1996, 1999; Roy et al. 2000). As would be expected for proteins that recycle between the ER and the cis Golgi P24 family members (p24A, gp25L2, and TMP21) ERGIC-53, Rab1a, Sec22b, and the KDEL receptor were detected in smooth microsomes (Gilchrist et al. 2006) in ERGIC (Breuza et al. 2004) and in ER/Golgi-derived vesicles marked by the cis Golgi marker p115 (Foster et al. 2006). The tER plays a key role in the formation of cargo exit sites and since the extent and amount of cargo exit sites may vary between different cell types (Fan et al. 2003; Bannykh et al. 1996) the relative content of cargo and membrane traffic proteins is expected to vary in proteomics studies using membrane-derivatives of the tER from different cell types.
ER chaperones
The ER functions to both co- and post-translationally modify newly synthesized proteins and lipids and sort them for housekeeping within the ER and for transport to their sites of function away from the ER (Palade 1975). A variety of proteins both membrane-bound and luminal are involved in the control of maturation of nascent proteins and lipids and include proteins involved in folding, in the regulation of calcium homeostasis, in oligomerization, and glycosylation (Hebert and Molinari 2007). Many such proteins have been detected by proteomics analysis of fractions representing different domains of the ER. Knoblach et al. (2003) have concentrated their efforts on the analysis of luminal proteins in purified rough microsomes from mouse liver and have detected BiP, GRP94, lectin-like chaperones (calnexin, calreticulin), peptidylprolyl isomerases, thiol disulphide oxidoreductases (PDI, P5 (CaBP1), ERp72, ERp57, ERp44, ERp29, and ERp46). Quantitative proteomics revealed these same chaperones to be in similar concentrations in rough and smooth microsomes from rat liver (Gilchrist et al. 2006). Detergent extraction of the rough and smooth microsomes after salt wash revealed the presence of the ER chaperones in the aqueous phase of the detergent extracts, thus confirming the luminal localization for all these proteins (Gilchrist et al. 2006).
The subunits of the chaperonin containing TCP-1 were detected associated with ER subcellular fractions (Foster et al. 2006; Gilchrist et al. 2006). Although tubulin and actin are well-known substrates for chaperonin t-complex proteins, new substrates are being defined and include proteins involved in cell cycle events (Liu et al. 2005). A role for chaperonin containing TCP-1 in protecting proteolytic intermediates in the MHC class I antigen-processing pathway has been described (Kunisawa and Shastri 2003). Whether this chaperone complex participates in MHC class I antigen processing at the cytosolic surface of a specific ER subcompartment remains to be confirmed.
Calcium-handling proteins
The ER plays an important role in Ca2+-homeostasis. This organelle has been described as a heterogeneous compartment with respect to the distribution of its Ca2+-handling proteins including the Ca2+-binding proteins, the Ca2+-pumps, and the Ca2+-release channels (Papp et al. 2003). Whereas the quantitative proteomics analysis of Gilchrist et al. (2006) revealed the Ca2+-pump SERCA2 and the Ca2+-release channel inositol 1,4,5-trisphosphate (InsP3) receptor to be enriched in smooth microsomes most Ca2+-binding proteins including calreticulin, glucose-regulated protein 78 and 94 (Grp78/BiP and Grp94), and protein disulfide isomerase, were similarly distributed between rough and smooth microsomes. Electron energy loss imaging analysis confirmed the heterogeneity of calcium distribution in ER of PC12 cells but was unable to distinguish distributions with respect to rough and smooth ER domains (Pezzati et al. 1997). The relative capacity for calcium storage and release of the rough and the smooth domains of the ER may vary between different cell types and the volume of these two subcompartments may influence this capacity.
Enzymes of lipid and glucose metabolism
The endoplasmic reticulum plays an important role in lipid biosynthesis and in glucose metabolism. Because of the large amount of sER in steroid-producing cells, lipid biosynthesis has often been attributed to the smooth domain of the ER. Indeed enzymes involved in lipid biosynthesis have been localized to the sER in such cells (Frederiks et al. 2007). However, smooth microsomes purified from the steroid-producing adrenocortical cells revealed not only enzymes involved in lipid synthesis but surprising high levels of translocation apparatus, and oligosaccharyltransferase complex proteins (Black et al. 2005). The implication of this finding is that lipogenic enzymes and other proteins involved in lipid metabolism should be easily recognized in both sER and rER domains. Lipogenic enzymes (ex. fatty acid synthase, acetyl-CoA carboxylase 1, and ATP citrate lyase) and lipid transfer proteins (ex. microsomal triacylglycerol transfer protein) indeed have been detected by proteomics analysis in both rough microsomal (Foster et al. 2006; Gilchrist et al. 2006; Knoblach et al. 2003) and smooth microsomal (Gilchrist et al. 2006) fractions. In terms of the distribution of proteins involved in lipid metabolism the data suggests a lack of distinction between sER and rER membrane domains. This suggestion is consistent with the previous morphological demonstration of lipid droplet formation in association with rER domains in situ (Robenek et al. 2006) and the demonstration of the formation of lipid droplets using ribosome-depleted rough microsomes in a cell-free reconstitution system (Paiement et al. 1994).
A number of proteins involved in the pentose-phosphate pathway (PPP) have been reported in association with ER subfractions including transketolase (Foster et al. 2006; Gilchrist et al. 2006), glucose-6-phosphate dehydrogenase (G6PD) (Gilchrist et al. 2006) and transaldolase (Gilchrist et al. 2006) following analysis by mass spectrometry. The detection of the transketolase and G6PD in the ER by mass spectrometry is consistent with previous electron microscope cytochemical studies showing these proteins in association with the ER in situ (Boren et al. 2006; Frederiks and Vreeling-Sindelarova 2001). Thus PPP enzymes may play a role in glucose metabolism at the level of the ER in specific cell types.
Proteins of detoxification and drug protein targets of the ER
The xenobiotic metabolism pathway of the ER includes functioning cytochrome p450 family (CYPs) proteins, UDP- glucuronosyltransferases, and carboxylesterases. Extensive biochemical and cytochemical data exist showing enrichment of such proteins in ER (reviewed in Seliskar and Rozman 2007) but little data differentiates between the rER and sER subcompartments. In the quantitative proteomics study of Gilchrist et al. (2006) over 30 different cytochrome P450s were detected as well as nine carboxylesterases and over 10 different glucuronosyltransferases and most of these proteins were in similar concentrations in rough and smooth microsomes. Continuity between the rough and smooth ER domains may endow specific ER proteins with the capacity to diffuse freely along the membranes (Nikonov et al. 2002) and within the lumen (Snapp et al. 2006), thus explaining equal distribution of enzymes involved in xenobiotic metabolism in rER and sER domains.
Endoplasmic reticulum proteins of detoxification and drug protein targets are now being examined quantitatively by proteomics in diseased states and under controlled drug treatments. Proteomics studies of liver microsomes under controlled drug treatments have revealed differential expression of specific ER proteins. Cytochromes P450 2B1, and 2B2, protein disulfide isomerase A3 and A6, and 78-kDa glucose-regulated protein were differentially expressed following phenobarbitol treatment (Galeva and Altermann 2002). p450 family proteins CYP1A2, −2A4/5, −2B10, −2B20, −2C29, −2C37, −2C38, −3A11, and −39A1 were up-regulated, and CYP2C40, −2E1, −3A41, and −27A1 were down-regulated by treatment with 1,4-bis-2-(3,5-dichloropyridyloxybenzene) (Lane et al. 2007).
Ubiquitin metabolizing enzymes
Endoplasmic reticulum-associated degradation (ERAD) is a regulated process whereby misfolded and unassembled proteins are recognized and retained in the ER by the quality-control apparatus and subsequently extracted, polyubiquitinated, and finally degraded in the cytoplasm by the multi-subunit 26S proteasome complex (reviewed by Meusser et al. 2005 and Romisch 2005). Ubiquitin and ubiquitin-activating enzymes were shown by electron microscope immuno-gold labeling to be associated with the post-ER/pre-Golgi network consisting of anastomosing tubulated and fenestrated membranes (Raposo et al. 1995). More recently misfolded protein was detected in an ER subdomain and in enlarged pre-Golgi intermediates by electron microscope immuno-gold labeling (Zuber et al. 2004). The quantitative proteomics analysis of Gilchrist et al. (2006) revealed proteasome subunits as well as ubiquitin metabolizing enzymes only in smooth microsomes. This data is consistent with a previous report showing that proteasomes were associated with the smooth endoplasmic reticulum and that they were practically absent from the rough endoplasmic reticulum (Palmer et al. 1996). Smooth microsomes used in the study of Gilchrist et al. (2006) may correspond to the compartment identified and described by Raposo et al. (1995) and Zuber et al. (2004). The smooth ER could be an important subcellular site for proteasome-dependent degradation of misfolded protein?
Proteins involved in antigen processing
Major histocompatibility complex (MHC) class I molecules present short, perfectly cleaved peptides on the cell surface for immune surveillance by T lymphocytes (Hammer et al. 2007). The intracellular processes that take part in creating the antigen-presenting machinery of the plasma membrane occur at the level of the ER and involve protein degradation, membrane translocation, and protein folding (reviewed by Elliott 2006 and Hammer et al. 2007). Protein degradation is carried out on the cytosolic side of the ER by the proteasome and the enzyme tripeptidyl peptidase II. The group II chaperonin TRiC (TCP-1 ring complex) is thought to chaperone the peptide between proteases and TAP the peptide transporter of the ER. TAP is composed of two transmembrane proteins, TAP1 and TAP2, and participates in the translocation of peptide to the lumen of the ER in an ATP-dependent step. On the luminal side of the ER the peptide is further processed by ERAAP, the ER aminopeptidase associated with antigen processing. The multicomponent MHC class I peptide-loading complex (PLC) includes MHC class I molecules, beta 2 microglobulin, the chaperone calreticulin, the oxidoreductases ERp57, and protein disulfide isomerase, the class I-specific accessory molecule tapasin and the peptide transporter TAP. This complex ensures the establishment of proper conformation of MHC class I molecules for peptide loading in the ER. A number of the components of the PLC have been reported in proteomics studies of ER fractions (Foster et al. 2006; Gilchrist et al. 2006). The study of Gilchrist et al. (2006) revealed higher amounts of beta 2 microglobulin, tapasin, and ERAAP in smooth microsomes compared to rough microsomes. Electron microscope immuno-gold labeling has previously revealed Class I molecules and beta 2 microglobulin in association with the post-ER/pre-Golgi network consisting of anastomosing tubulated and fenestrated membranes (Raposo et al. 1995). Whether the transitional zone of the ER, which contains the smooth ER is a major site for antigen processing and peptide loading onto the PLC remains to be confirmed.
Endoplasmic reticulum proteins were previously detected in purified phagosomes using proteomics analysis (Garin et al. 2001). This observation prompted consideration that the ER maybe involved in phagocytosis and indeed electron microscope cytochemical, electron microscope immuno-gold labeling, and biochemical data were obtained to testify to this possibility (Gagnon et al. 2002). Endoplasmic reticulum involvement in phagocytosis has been confirmed independently by other laboratories (Ackerman et al. 2006; Becker et al. 2005) and the findings have led to additional proteomics studies and the important conclusions explaining antigen cross-presentation (Guermonprez et al. 2003; Houde et al. 2003). Therefore even though organelle proteomics studies may often suggest the presence of organelle contaminants using intuitive reasoning and corollary experiments as was done by Michel Desjardins and colleagues, in the case of ER proteins associated with phagosomes such studies may also provide information leading to new paradigms.
Cytoskeletal proteins
The ER interacts with the cytoskeleton to maintain its position within sedentary cells and to establish new intracellular positions in motile cells. The ER contains a receptor for specific interaction with microtubules (Farah et al. 2005; Klopfenstein et al. 1998) and can move along both microtubules and actin cables (Allan and Vale 1994; Kachar and Reese 1988; Terasaki and Reese 1994). Based on these reports one might expect proteomics studies of the ER to reveal the presence of associated cytoskeletal proteins. Indeed, protein components of microtubules but not of microfilaments have previously been detected in association with ER subfractions from mammalian liver (Foster et al. 2006; Gilchrist et al. 2006). In addition a number of molecular motors including protein members of the myosin, kinesin, and dynein protein families have been detected in association with these same membrane fractions. Subcellular membrane movements are directed by molecular motors including myosin, dynein, and kinesin family molecular motors (Mallik and Gross 2004). Other cytoskeletal proteins, which have previously been detected in ER fractions include filamin A and B, the actin-binding proteins profilin 1, and transgelin 2.
Filamin A and B were detected in smooth ER fractions (Gilchrist et al. 2006). Filamin A has been implicated in intracellular traffic of several cell-surface receptors and was observed to bind one of these at the level of the endoplasmic reticulum (Feng et al. 2005; Liu et al. 1997). Therefore filamin A may bind yet unidentified nascent receptors in the ER in hepatocytes and promote traffic of these molecules to the cell surface.
The actin-binding proteins profilin 1 and transgelin 2 were detected in the ER fractions. Higher amounts of these proteins were found in association with smooth microsomes compared with that in rough microsomes (Gilchrist et al. 2006). Profilin 1 is able to bind membrane phosphoinositides and thus affect cell signaling and has been implicated in membrane traffic (Witke 2004), thus it maybe involved in ER membrane traffic in hepatocytes. Transgelin 2 mRNA was reported overexpressed in hepatocellular carcinoma and this protein has been proposed as a potential diagnostic marker for this disease (Shi et al. 2005). The role of transgelin 2 at the level of the ER remains to be determined.
Cytoskeleton-associated protein 4/CLIMP-63 was reported predominantly in rough microsomes of rat liver by the studies of Gilchrist et al. 2006. CLIMP-63 serves as a specific ER- microtubule receptor (Klopfenstein et al. 1998) and was shown to link the ER to microtubules via the neuronal microtubule-associated protein 2 (MAP-2) in the dendritic compartment of neurons (Farah et al. 2005). Interestingly, the non-neuronal microtubule-associated protein 4 (MAP-4), which has an identical microtubule-binding domain to MAP-2 was also detected in association with rough microsomes (Gilchrist et al. 2006). Whether MAP-4 and CLIMP-63 make a link between the rER and microtubules thus immobilizing rER structure in hepatocytes remains to be confirmed.
Reticulons constitute a family of proteins implicated in the tubular structure of the smooth ER (Voeltz et al. 2006). Reticulon 3 was detected by mass spectrometry in smooth microsomes of rat liver by Gilchrist et al. (2006) and over-expression of this protein in Hela cells revealed production of fluorescent tubules, which co-distributed along microtubules as revealed by double-labeling immunofluorescence (Wakana et al. 2005). As proposed by Shibata et al. (2006) reticulons promote long unbranched ER tubules. Endoplasmic reticulum tubules have previously been shown to be highly dynamic structures capable of moving along tracks of microtubules (Waterman-Storer and Salmon 1998). Perhaps the tubular structure of the ER favors ER/motor/microtubule interaction? However if reticulons promote long unbranched ER tubules, how do they affect tritubular jonctions as observed in the large inter-connecting networks of tubules in sER in steroid cells or in hepatocytes?
Proteins involved in signaling
The ER is involved in many signaling pathways including the unfolded protein response (UPR) (Bernales et al. 2006), apoptosis (Chae et al. 2004; Nakajima et al. 2004), ubiquitination and proteasome degradation (Kostova and Wolf 2003) and some of the involved signaling molecules have been shown to associate in a transient manner with ER membranes. This includes Ras protein, which is involved in cell signaling (Sobering et al. 2004), the AAA ATPase p97, which is involved in ER assembly (Lavoie et al. 2000) and in proteasome degradation (Ye et al. 2003) and BAX, which is involved in apoptosis (Zong et al. 2003). Excluding proteins involved in ubiquitination and proteasome degradation (see above), few signaling proteins have yet to be detected in proteomics studies of ER fractions. This is true for a number of regulatory proteins such as SREBP-1, which regulates lipogenic enzymes (Bengoechea-Alonso and Ericsson 2007), Ire-1 which regulates the UPR (Bernales et al. 2006) and Bax, which is involved in apoptosis (Hetz et al. 2006). These signaling proteins probably represent low abundance proteins of the ER and require up-regulation following physiological activation to be detected by existing mass spectrometry techniques.
Under conditions of accumulation of misfolded and unassembled proteins, the UPR acts to modify the ER by up-regulating chaperones so as to increase its folding capacity. Morand et al. (2005) used quantitative proteomics analysis and revealed a number of chaperones (GRP94 and PDI) to be up-regulated in ER microsomes from the liver of a fructose-fed, insulin-resistant hamster model and up-regulation was attributed to ER stress. Ras protein and p97 have been reported in association with ER fractions (Gilchrist et al. 2006; Galeva and Altermann 2002). Association of Ras with the ER is consistent with previous reports of the association of Ras with membranes of the secretory pathway including ER (Zheng et al. 2007). Association of p97 with ER is consistent with its role in ER assembly (Lavoie et al. 2000) and proteasome degradation (Ye et al. 2003).
Phosphoproteins associated with the ER
The ER is a membrane-bound compartment that functions by interacting with cytoplasmic and luminal-soluble proteins. Among the soluble proteins that interact with the ER membrane in a transient manner are signaling proteins. Reversible phosphorylation has been shown to control protein interaction at the ER of several signaling proteins including valosin-containing protein/p97 (Lavoie et al. 2000) BCL-2 (Lin et al. 2006) and Ire1 (Bernales et al. 2006). In addition, reversible phosphorylation has been shown to play a role in the control of ER structure. For example, phosphorylation of CLIMP-63 was shown to affect ER structure during interphase (Vedrenne et al. 2005) and phosphorylation of p47, a cofactor of p97, was shown to control ER structure at mitosis (Kano et al. 2005). Since the ER controls key metabolic events including events crucial to cell survival, it is expected that new phosphoproteins are yet to be identified, especially in relations to control of cell cycle events.
A number of proteins have been detected in ER fractions, which have previously been shown to be phosphoproteins. Phosphoproteins were identified in transformed cells by analysis of tyrosine-phosphorylated peptides immunoprecipitated using anti-phosphotyrosine antibodies (Rush et al. 2005). A significant number of these previously identified tyrosine-phosphorylated proteins have in fact been detected in association with ER membranes by proteomics analysis (Foster et al. 2006; Gilchrist et al. 2006). Many of these proteins are not conventional ER proteins but rather are better known as cytosolic proteins; these include proteins involved in mRNA metabolism (PABP1, YB-1, ELAV-like 1, eEF-1-alpha), proteins involved in phospholipid binding (annexin I, annexin II), and proteins involved in de novo lipid synthesis (ATP citrate lyase). Some of these proteins are substrates of key signaling pathways including the Akt-signaling pathway (ex. ATP citrate lyase, Berwick et al. 2002) and some have been shown to have a phosphorylation status which has been associated with subcellular trafficking (Hibino et al. 2006). Thus proteins previously defined as tyrosine-phosphorylated proteins were observed in ER membranes by proteomics analysis and these may undergo cycles of tyrosine phosphorylation and dephosphorylation promoting specific ER-related functions in cells, which are yet to be better defined.
Serine/threonine protein phosphorylation can also modify subcellular protein localization. Phosphorylation of the disc’s large tumor suppressor protein controls its membrane localization (Massimi et al. 2006). CLIMP-63 is serine phosphorylated during mitosis and regulates ER-microtubule binding (Vedrenne et al. 2005).
Conclusions and future perspectives
From available proteomics data parts of which are supported by electron microscope protein localization studies some tentative conclusions can be drawn about the relative segregation of proteins and molecular machines in the subcompartments of the ER. CLIMP-63 and ribosomes are enriched in rER. Reticulon, enzymes involved in ubiquitination, the proteasome, some cytoskeletal proteins, proteins involved in antigen processing, and coat proteins are enriched in sER. In contrast, proteins that appear equally distributed between rER and sER include proteins of the translocon, biosynthetic cargo, chaperones, proteins of detoxification, and proteins involved in lipid and glucose metabolism.
Because proteomics of ER subcompartments is carried out using subcellular fractions, there are limitations that have to be considered when trying to interpret the results of the protein analysis. For example, the relative purity of the fractions will vary and proteins of fragments of contaminating organelles will be present in the fraction and be identified in the analysis. Moreover molecular dynamics at the cytosolic and luminal surfaces of the ER have to be taken into consideration. The composition of the ER is subject to change based on molecular interactions occurring on both cytosolic and luminal sides of the ER membrane. On the cytosolic side of the ER interaction with the cytoskeleton (to change the location, shape or size of the ER) or interaction with signaling proteins (to activate specific signaling pathways, e.g., apoptosis) leads to transient associations between the ER and cytosolic proteins. On the luminal side of the ER interactions between newly synthesized proteins and chaperones can vary under specific conditions (for example during ER stress) and will affect the overall molecular composition. Up-regulation of proteins involved in detoxification may occur under exposure to toxic chemicals and affect ER composition. Thus depending on the cell physiology protein associations with the ER may vary. Dynamics of protein interactions in organelles are often controlled by posttranslational modifications including phosphorylation. Understanding such modifications is key to understanding site-specific protein function.
Proteomics studies of the subcompartments of the ER have lead to insights into the function of the different compartments of this organelle and new paradigms. However data obtained using proteomics analysis should be complimented by cytological techniques to confirm the localization of the proteins in the ER subcompartments and molecular biology should be used to modulate protein expression to examine the function. The combination of these approaches not only will yield new information about the proteins but they will also expand knowledge on the protein families to which they belong and/or protein complexes of which they are part of.
References
Ackerman AL, Giodini A, Cresswell P (2006) A role for the endoplasmic reticulum protein retrotranslocation machinery during crosspresentation by dendritic cells. Immunity 25:607–617
Allan V, Vale R (1994) Movement of membrane tubules along microtubules in vitro: evidence for specialised sites of motor attachment. J Cell Sci 107(Pt 7):1885–1897
Bannykh SI, Rowe T, Balch WE (1996) The organization of endoplasmic reticulum export complexes. J Cell Biol 135:19–35
Baumann O, Walz B (2001) Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int Rev Cytol 205:149–214
Becker T, Volchuk A, Rothman JE (2005) Differential use of endoplasmic reticulum membrane for phagocytosis in J774 macrophages. Proc Natl Acad Sci USA 102:4022–4026
Bengoechea-Alonso MT, Ericsson J (2007) SREBP in signal transduction: cholesterol metabolism and beyond. Curr Opin Cell Biol 19:215–222
Bernales S, Papa FR, Walter P (2006) Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol 22:487–508
Berwick DC, Hers I, Heesom KJ, Moule SK, Tavare JM (2002) The identification of ATP-citrate lyase as a protein kinase B (Akt) substrate in primary adipocytes. J Biol Chem 277:33895–33900
Black VH, Sanjay A, van Leyen K, Lauring B, Kreibich G (2005) Cholesterol and steroid synthesizing smooth endoplasmic reticulum of adrenocortical cells contains high levels of proteins associated with the translocation channel. Endocrinology 146:4234–4249
Boren J, Ramos-Montoya A, Bosch KS, Vreeling H, Jonker A, Centelles JJ, Cascante M, Frederiks WM (2006) In situ localization of transketolase activity in epithelial cells of different rat tissues and subcellularly in liver parenchymal cells. J Histochem Cytochem 54:191–199
Breuza L, Halbeisen R, Jeno P, Otte S, Barlowe C, Hong W, Hauri HP (2004) Proteomics of endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membranes from brefeldin A-treated HepG2 cells identifies ERGIC-32, a new cycling protein that interacts with human Erv46. J Biol Chem 279:47242–47253
Chae HJ, Kim HR, Xu C, Bailly-Maitre B, Krajewska M, Krajewski S, Banares S, Cui J, Digicaylioglu M, Ke N, Kitada S, Monosov E, Thomas M, Kress CL, Babendure JR, Tsien RY, Lipton SA, Reed JC (2004) BI-1 regulates an apoptosis pathway linked to endoplasmic reticulum stress. Mol Cell 15:355–366
Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ (2002) Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol 158:915–927
Dreger M, Bengtsson L, Schoneberg T, Otto H, Hucho F (2001) Nuclear envelope proteomics: novel integral membrane proteins of the inner nuclear membrane. Proc Natl Acad Sci USA 98:11943–11948
Dreyfuss G, Kim VN, Kataoka N (2002) Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 3:195–205
Ellgaard L, Molinari M, Helenius A (1999) Setting the standards: quality control in the secretory pathway. Science 286:1882–1888
Elliott T (2006) The ‘chop-and-change’ of MHC class I assembly. Nat Immunol 7:7–9
Fan JY, Roth J, Zuber C (2003) Ultrastructural analysis of transitional endoplasmic reticulum and pre-Golgi intermediates: a highway for cars and trucks. Histochem Cell Biol 120:455–463
Farah CA, Liazoghli D, Perreault S, Desjardins M, Guimont A, Anton A, Lauzon M, Kreibich G, Paiement J, Leclerc N (2005) Interaction of microtubule-associated protein-2 and p63: a new link between microtubules and rough endoplasmic reticulum membranes in neurons. J Biol Chem 280:9439–9449
Feng S, Lu X, Kroll MH (2005) Filamin A binding stabilizes nascent glycoprotein Ibalpha trafficking and thereby enhances its surface expression. J Biol Chem 280:6709–6715
Foster LJ, de Hoog CL, Zhang Y, Zhang Y, Xie X, Mootha VK, Mann M (2006) A mammalian organelle map by protein correlation profiling. Cell 125:187–199
Frederiks WM, Kummerlin IP, Bosch KS, Vreeling-Sindelarova H, Jonker A, Van Noorden CJ (2007) NADPH production by the pentose phosphate pathway in the zona fasciculata of rat adrenal gland. J Histochem Cytochem 55:975–980
Frederiks WM, Vreeling-Sindelarova H (2001) Localization of glucose-6-phosphate dehydrogenase activity on ribosomes of granular endoplasmic reticulum, in peroxisomes and peripheral cytoplasm of rat liver parenchymal cells. Histochem J 33:345–353
Gagnon E, Duclos S, Rondeau C, Chevet E, Cameron PH, Steele-Mortimer O, Paiement J, Bergeron JJ, Desjardins M (2002) Endoplasmic reticulum-mediated phagocytosis is a mechanism of entry into macrophages. Cell 110:119–131
Galeva N, Altermann M (2002) Comparison of one-dimensional and two-dimensional gel electrophoresis as a separation tool for proteomic analysis of rat liver microsomes: cytochromes P450 and other membrane proteins. Proteomics 2:713–722
Garin J, Diez R, Kieffer S, Dermine JF, Duclos S, Gagnon E, Sadoul R, Rondeau C, Desjardins M (2001) The phagosome proteome: insight into phagosome functions. J Cell Biol 152:165–180
Gilchrist A, Au CE, Hiding J, Bell AW, Fernandez-Rodriguez J, Lesimple S, Nagaya H, Roy L, Gosline SJ, Hallett M, Paiement J, Kearney RE, Nilsson T, Bergeron JJ (2006) Quantitative proteomics analysis of the secretory pathway. Cell 127:1265–1281
Gilmore R (1993) Protein translocation across the endoplasmic reticulum: a tunnel with toll booths at entry and exit. Cell 75:589–592
Guermonprez P, Saveanu L, Kleijmeer M, Davoust J, Van Endert P, Amigorena S (2003) ER-phagosome fusion defines an MHC class I cross-presentation compartment in dendritic cells. Nature 425:397–402
Hammer GE, Kanaseki T, Shastri N (2007) The final touches make perfect the peptide-MHC class I repertoire. Immunity 26:397–406
Hauri HP, Kappeler F, Andersson H, Appenzeller C (2000) ERGIC-53 and traffic in the secretory pathway. J Cell Sci 113(Pt 4):587–596
Hebert DN, Molinari M (2007) In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol Rev 87:1377–1408
Hetz C, Bernasconi P, Fisher J, Lee AH, Bassik MC, Antonsson B, Brandt GS, Iwakoshi NN, Schinzel A, Glimcher LH, Korsmeyer SJ (2006) Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science 312:572–576
Hibino Y, Usui T, Morita Y, Hirose N, Okazaki M, Sugano N, Hiraga K (2006) Molecular properties and intracellular localization of rat liver nuclear scaffold protein P130. Biochim Biophys Acta 1759:195–207
Houde M, Bertholet S, Gagnon E, Brunet S, Goyette G, Laplante A, Princiotta MF, Thibault P, Sacks D, Desjardins M (2003) Phagosomes are competent organelles for antigen cross-presentation. Nature 425:402–406
Johnson AE, van Waes MA (1999) The translocon: a dynamic gateway at the ER membrane. Annu Rev Cell Dev Biol 15:799–842
Kachar B, Reese TS (1988) The mechanism of cytoplasmic streaming in characean algal cells: sliding of endoplasmic reticulum along actin filaments. J Cell Biol 106:1545–1552
Kano F, Kondo H, Yamamoto A, Tanaka AR, Hosokawa N, Nagata K, Murata M (2005) The maintenance of the endoplasmic reticulum network is regulated by p47, a cofactor of p97, through phosphorylation by cdc2 kinase. Genes Cells 10:333–344
Klopfenstein DR, Kappeler F, Hauri HP (1998) A novel direct interaction of endoplasmic reticulum with microtubules. Embo J 17:6168–6177
Knoblach B, Keller BO, Groenendyk J, Aldred S, Zheng J, Lemire BD, Li L, Michalak M (2003) ERp19 and ERp46, new members of the thioredoxin family of endoplasmic reticulum proteins. Mol Cell Proteomics 2:1104–1119
Kostova Z, Wolf DH (2003) For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. Embo J 22:2309–2317
Kunisawa J, Shastri N (2003) The group II chaperonin TRiC protects proteolytic intermediates from degradation in the MHC class I antigen processing pathway. Mol Cell 12:565–576
Lane CS, Wang Y, Betts R, Griffiths WJ, Patterson LH (2007) Comparative cytochrome P450 proteomics in the livers of immunodeficient mice using 18O stable isotope labeling. Mol Cell Proteomics 6:953–962
Lavoie C, Lanoix J, Kan FW, Paiement J (1996) Cell-free assembly of rough and smooth endoplasmic reticulum. J Cell Sci 109(Pt 6):1415–1425
Lavoie C, Paiement J, Dominguez M, Roy L, Dahan S, Gushue JN, Bergeron JJ (1999) Roles for alpha(2)p24 and COPI in endoplasmic reticulum cargo exit site formation. J Cell Biol 146:285–299
Lavoie C, Chevet E, Roy L, Tonks NK, Fazel A, Posner BI, Paiement J, Bergeron JJ (2000) Tyrosine phosphorylation of p97 regulates transitional endoplasmic reticulum assembly in vitro. Proc Natl Acad Sci USA 97:13637–13642
Lee MC, Miller EA, Goldberg J, Orci L, Schekman R (2004) Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol 20:87–123
Lerner RS, Nicchitta CV (2006) mRNA translation is compartmentalized to the endoplasmic reticulum following physiological inhibition of cap-dependent translation. Rna 12:775–789
Lin SS, Bassik MC, Suh H, Nishino M, Arroyo JD, Hahn WC, Korsmeyer SJ, Roberts TM (2006) PP2A regulates BCL-2 phosphorylation and proteasome-mediated degradation at the endoplasmic reticulum. J Biol Chem 281:23003–23012
Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR 3rd (1999) Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol 17:676–682
Liu G, Thomas L, Warren RA, Enns CA, Cunningham CC, Hartwig JH, Thomas G (1997) Cytoskeletal protein ABP-280 directs the intracellular trafficking of furin and modulates proprotein processing in the endocytic pathway. J Cell Biol 139:1719–1733
Liu X, Lin CY, Lei M, Yan S, Zhou T, Erikson RL (2005) CCT chaperonin complex is required for the biogenesis of functional Plk1. Mol Cell Biol 25:4993–5010
Mallik R, Gross SP (2004) Molecular motors: strategies to get along. Curr Biol 14:R971–R982
Massimi P, Narayan N, Cuenda A, Banks L (2006) Phosphorylation of the discs large tumour suppressor protein controls its membrane localisation and enhances its susceptibility to HPV E6-induced degradation. Oncogene 25:4276–4285
Meusser B, Hirsch C, Jarosch E, Sommer T (2005) ERAD: the long road to destruction. Nat Cell Biol 7:766–772
Morand JP, Macri J, Adeli K (2005) Proteomic profiling of hepatic endoplasmic reticulum-associated proteins in an animal model of insulin resistance and metabolic dyslipidemia. J Biol Chem 280:17626–17633
Nakajima K, Hirose H, Taniguchi M, Kurashina H, Arasaki K, Nagahama M, Tani K, Yamamoto A, Tagaya M (2004) Involvement of BNIP1 in apoptosis and endoplasmic reticulum membrane fusion. Embo J 23:3216–3226
Nikonov AV, Snapp E, Lippincott-Schwartz J, Kreibich G (2002) Active translocon complexes labeled with GFP-Dad1 diffuse slowly as large polysome arrays in the endoplasmic reticulum. J Cell Biol 158:497–506
Nikonov AV, Hauri HP, Lauring B, Kreibich G (2007) Climp-63-mediated binding of microtubules to the ER affects the lateral mobility of translocon complexes. J Cell Sci 120:2248–2258
Nilsson I, Kelleher DJ, Miao Y, Shao Y, Kreibich G, Gilmore R, von Heijne G, Johnson AE (2003) Photocross-linking of nascent chains to the STT3 subunit of the oligosaccharyltransferase complex. J Cell Biol 161:715–725
Nilsson J, Sengupta J, Frank J, Nissen P (2004) Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep 5:1137–1141
Osborne AR, Rapoport TA, van den Berg B (2005) Protein translocation by the Sec61/SecY channel. Annu Rev Cell Dev Biol 21:529–550
Paiement J, Lavoie C, Gavino GR, Gavino VC (1994) Modulation of GTP-dependent fusion by linoleic and arachidonic acid in derivatives of rough endoplasmic reticulum from rat liver. Biochim Biophys Acta 1190:199–212
Palade G (1975) Intracellular aspects of the process of protein synthesis. Science 189:347–358
Palmer A, Rivett AJ, Thomson S, Hendil KB, Butcher GW, Fuertes G, Knecht E (1996) Subpopulations of proteasomes in rat liver nuclei, microsomes and cytosol. Biochem J 316(Pt 2):401–407
Papp S, Dziak E, Michalak M, Opas M (2003) Is all of the endoplasmic reticulum created equal? The effects of the heterogeneous distribution of endoplasmic reticulum Ca2+-handling proteins. J Cell Biol 160:475–479
Pezzati R, Bossi M, Podini P, Meldolesi J, Grohovaz F (1997) High-resolution calcium mapping of the endoplasmic reticulum-golgi-exocytic membrane system. Electron energy loss imaging analysis of quick frozen-freeze dried PC12 cells. Mol Biol Cell 8:1501–1512
Rapoport TA, Jungnickel B, Kutay U (1996) Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem 65:271–303
Raposo G, van Santen HM, Leijendekker R, Geuze HJ, Ploegh HL (1995) Misfolded major histocompatibility complex class I molecules accumulate in an expanded ER-golgi intermediate compartment. J Cell Biol 131:1403–1419
Robenek H, Hofnagel O, Buers I, Robenek MJ, Troyer D, Severs NJ (2006) Adipophilin-enriched domains in the ER membrane are sites of lipid droplet biogenesis. J Cell Sci 119:4215–4224
Romisch K (2005) Endoplasmic reticulum-associated degradation. Annu Rev Cell Dev Biol 21:435–456
Roth J, Zuber C, Guhl B, Fan JY, Ziak M (2002) The importance of trimming reactions on asparagine-linked oligosaccharides for protein quality control. Histochem Cell Biol 117:159–169
Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT (2000) The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol 148:635–651
Roy L, Bergeron JJ, Lavoie C, Hendriks R, Gushue J, Fazel A, Pelletier A, Morre DJ, Subramaniam VN, Hong W, Paiement J (2000) Role of p97 and syntaxin 5 in the assembly of transitional endoplasmic reticulum. Mol Biol Cell 11:2529–2542
Ruddock LW, Molinari M (2006) N-glycan processing in ER quality control. J Cell Sci 119:4373–4380
Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ (2005) Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol 23:94–101
Saraste J, Kuismanen E (1992) Pathways of protein sorting and membrane traffic between the rough endoplasmic reticulum and the Golgi complex. Semin Cell Biol 3:343–355
Schirmer EC, Florens L, Guan T, Yates JR 3rd, Gerace L (2003) Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 301:1380–1382
Seliskar M, Rozman D (2007) Mammalian cytochromes P450–importance of tissue specificity. Biochim Biophys Acta 1770:458–466
Shi YY, Wang HC, Yin YH, Sun WS, Li Y, Zhang CQ, Wang Y, Wang S, Chen WF (2005) Identification and analysis of tumour-associated antigens in hepatocellular carcinoma. Br J Cancer 92:929–934
Shibata Y, Voeltz GK, Rapoport TA (2006) Rough sheets and smooth tubules. Cell 126:435–439
Shibatani T, David LL, McCormack AL, Frueh K, Skach WR (2005) Proteomic analysis of mammalian oligosaccharyltransferase reveals multiple subcomplexes that contain Sec61, TRAP, and two potential new subunits. Biochemistry 44:5982–5992
Shyu AB, Wilkinson MF (2000) The double lives of shuttling mRNA binding proteins. Cell 102:135–138
Snapp EL, Sharma A, Lippincott-Schwartz J, Hegde RS (2006) Monitoring chaperone engagement of substrates in the endoplasmic reticulum of live cells. Proc Natl Acad Sci USA 103:6536–6541
Sobering AK, Watanabe R, Romeo MJ, Yan BC, Specht CA, Orlean P, Riezman H, Levin DE (2004) Yeast Ras regulates the complex that catalyzes the first step in GPI-anchor biosynthesis at the ER. Cell 117:637–648
Terasaki M, Reese TS (1994) Interactions among endoplasmic reticulum, microtubules, and retrograde movements of the cell surface. Cell Motil Cytoskeleton 29:291–300
Vedrenne C, Klopfenstein DR, Hauri HP (2005) Phosphorylation controls CLIMP-63-mediated anchoring of the endoplasmic reticulum to microtubules. Mol Biol Cell 16:1928–1937
Visa N, Alzhanova-Ericsson AT, Sun X, Kiseleva E, Bjorkroth B, Wurtz T, Daneholt B (1996) A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell 84:253–264
Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA (2006) A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124:573–586
Wakana Y, Koyama S, Nakajima K, Hatsuzawa K, Nagahama M, Tani K, Hauri HP, Melancon P, Tagaya M (2005) Reticulon 3 is involved in membrane trafficking between the endoplasmic reticulum and Golgi. Biochem Biophys Res Commun 334:1198–1205
Waterman-Storer CM, Salmon ED (1998) Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr Biol 8:798–806
Witke W (2004) The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol 14:461–469
Wool IG, Chan YL, Gluck A (1995) Structure and evolution of mammalian ribosomal proteins. Biochem Cell Biol 73:933–947
Ye Y, Meyer HH, Rapoport TA (2003) Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol 162:71–84
Zheng H, McKay J, Buss JE (2007) H-Ras does not need COP I- or COP II-dependent vesicular transport to reach the plasma membrane. J Biol Chem 282:25760–25768
Zong WX, Li C, Hatzivassiliou G, Lindsten T, Yu QC, Yuan J, Thompson CB (2003) Bax and Bak can localize to the endoplasmic reticulum to initiate apoptosis. J Cell Biol 162:59–69
Zuber C, Fan JY, Guhl B, Roth J (2004) Misfolded proinsulin accumulates in expanded pre-Golgi intermediates and endoplasmic reticulum subdomains in pancreatic beta cells of Akita mice. Faseb J 18:917–919
Acknowledgments
We would like to thank Nicole Leclerc for critical reading of the manuscript. Figure 1C is a courtesy of Christian Zuber and Jürgen Roth. C.L is supported by grants from the Canadian Institutes for Health Research and a Canada Research Chair.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Lavoie, C., Paiement, J. Topology of molecular machines of the endoplasmic reticulum: a compilation of proteomics and cytological data . Histochem Cell Biol 129, 117–128 (2008). https://doi.org/10.1007/s00418-007-0370-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-007-0370-y