Abstract
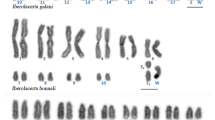
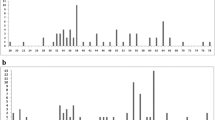
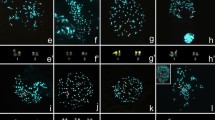
Cytogenetics has historically played a key role in research on squirrel monkey (genus Saimiri) evolutionary biology. Squirrel monkeys have a diploid number of 2n = 44, but vary in fundamental number (FN). Apparently, differences in FN have phylogenetic implications and are correlated with geographic regions. A number of hypothetical mechanisms were proposed to explain difference in FN: translocations, heterochromatin, or, most commonly, pericentric inversions. Recently, an additional mechanism, centromere repositioning, was discovered, which can alter chromosome morphology and FN. Here, we used chromosome banding, chromosome painting, and BAC-FISH to test these hypotheses. We demonstrate that centromere repositioning on chromosomes 5 and 15 is the mechanism that accounts for differences in FN. Current phylogenomic trees of platyrrhines provide a temporal framework for evolutionary new centromeres (ENC) in Saimiri. The X-chromosome ENC could be up to 15 million years (my) old that on chromosome 5 as recent as 0.3 my. The chromosome 15 ENC is intermediate, as young as 2.24 my. All ENC have abundant satellite DNAs indicating that the maturation process was fairly rapid. Callithrix jacchus was used as an outgroup for the BAC-FISH data analysis. Comparison with scaffolds from the S. boliviensis genome revealed an error in the last marmoset genome release. Future research including at the sequence level will provide better understanding of chromosome evolution in Saimiri and other platyrrhines. Probably other cases of differences in chromosome morphology and FN, both within and between taxa, will be shown to be due to centromere repositioning and not pericentric inversions.




Similar content being viewed by others
References
Cambefort Y, Moro F (1978) Cytogenetics and taxonomy of some South Bolivian monkeys. Folia Primatol (Basel) 29:307–314
Capozzi O, Archidiacono N, Lorusso N, Stanyon R, Rocchi M (2015) The 14/15 association as a paradigmatic example of tracing karyotype evolution in New World monkeys. Chromosoma
Carbone L, Nergadze SG, Magnani E, Misceo D, Francesca Cardone M, Roberto R, Bertoni L, Attolini C, Francesca Piras M, de Jong P, Raudsepp T, Chowdhary BP, Guerin G, Archidiacono N, Rocchi M, Giulotto E (2006) Evolutionary movement of centromeres in horse, donkey, and zebra. Genomics 87:777–782
Chen TR, Ruddle FH (1971) Karyotype analysis utilizing differentially stained constitutive heterochromatin of human and murine chromosomes. Chromosoma 34:51–72
Chiou KL, Pozzi L, Lynch Alfaro JW, Di Fiore A (2011) Pleistocene diversification of living squirrel monkeys (Saimiri spp.) inferred from complete mitochondrial genome sequences. Mol Phylogenet Evol 59:736–745
Craig-Holmes AP, Shaw MW (1971) Polymorphism of human constitutive heterochromatin. Science 174:702–704
Dumas F, Stanyon R, Sineo L, Stone G, Bigoni F (2007) Phylogenomics of species from four genera of New World monkeys by flow sorting and reciprocal chromosome painting. BMC Evol Biol 7(Suppl 2):S11
Dutrillaux B, Couturier J (1981) The ancestral karyotype of platyrrhine monkeys. Cytogenet Cell Genet 30:232–242
Hasson D, Alonso A, Cheung F, Tepperberg JH, Papenhausen PR, Engelen JJ, Warburton PE (2011) Formation of novel CENP-A domains on tandem repetitive DNA and across chromosome breakpoints on human chromosome 8q21 neocentromeres. Chromosoma 120:621–632
Jones TC, Thorington RW, Hu MM, Adams E, Cooper RW (1973) Karyotypes of squirrel monkeys (Saimiri sciureus) from different geographic regions. Am J Phys Anthropol 38:269–277
Lau YF, Arrighi FE (1976) Studies of the squirrel monkey, Saimiri sciureus, genome. I. Cytological characterization of chromosomal heterozygosity. Cytogenet Cell Genet 17:51–60
Lavergne A, Ruiz-Garcia M, Catzeflis F, Lacote S, Contamin H, Mercereau-Puijalon O, Lacoste V, de Thoisy B (2010) Phylogeny and phylogeography of squirrel monkeys (genus Saimiri) based on cytochrome b genetic analysis. Am J Primatol 72:242–253
Lynch Alfaro JW, Boubli JP, Paim FP, Ribas CC, Silva MN, Messias MR, Rohe F, Merces MP, Silva Junior JS, Silva CR, Pinho GM, Koshkarian G, Nguyen MT, Harada ML, Rabelo RM, Queiroz HL, Alfaro ME, Farias IP (2015) Biogeography of squirrel monkeys (genus Saimiri): south-Central Amazon origin and rapid pan-Amazonian diversification of a lowland primate. Mol Phylogenet Evol 82(Pt B):436–454
Ma NS, Jones TC (1975) Added heterochromatin segments in chromosomes of squirrel monkeys (Saimiri sciureus). Folia Primatol (Basel) 24:282–292
Marmoset Genome S, Analysis C (2014) The common marmoset genome provides insight into primate biology and evolution. Nat Genet 46:850–857
Moore CM, Harris CP, Abee CR (1990) Distribution of chromosomal polymorphisms in three subspecies of squirrel monkeys (genus Saimiri). Cytogenet Cell Genet 53:118–122
Oakenfull EA, Clegg JB (1998) Phylogenetic relationships within the genus Equus and the evolution of alpha and theta globin genes. J Mol Evol 47:772–783
Oakenfull EA, Lim HN, Ryder OA (2000) A survey of equid mitochondrial DNA: implications for the evolution, genetic diversity and conservation of Equus. Conserv Genet 1:341–355
Pardo-Manuel de Villena F, Sapienza C (2001) Transmission ratio distortion in offspring of heterozygous female carriers of Robertsonian translocations. Hum Genet 108:31–36
Perelman P, Johnson WE, Roos C, Seuanez HN, Horvath JE, Moreira MA, Kessing B, Pontius J, Roelke M, Rumpler Y, Schneider MP, Silva A, O’Brien SJ, Pecon-Slattery J (2011) A molecular phylogeny of living primates. PLoS Genet 7:e1001342
Rocchi M, Archidiacono N, Schempp W, Capozzi O, Stanyon R (2012) Centromere repositioning in mammals. Heredity (Edinb) 108:59–67
Ruiz-Garcia M, Luengas-Villamil K, Leguizamon N, de Thoisy B, Galvez H (2015) Molecular phylogenetics and phylogeography of all the Saimiri taxa (Cebidae, primates) inferred from mt COI and COII gene sequences. Primates; journal of primatology 56:145–161
Scammell JG, Wright JL, Tuck-Muller CM (2001) The origin of four squirrel monkey cell lines established by karyotype analysis. Cytogenet Cell Genet 93:263–264
Small MF, Stanyon R, Smith DG, Sinneo L (1985) High-resolution chromosomes of rhesus macaques (Macaca mulatta). Am J Primatol 9:63–67
Springer MS, Meredith RW, Gatesy J, Emerling CA, Park J, Rabosky DL, Stadler T, Steiner C, Ryder OA, Janečka JE, Fisher CA, Murphy WJ, Stanyon R (2012) Macroevolutionary dynamics and historical biogeography of primate diversification inferred from a species supermatrix. PLoS ONE 7(11):e49521. doi:10.1371/journal.pone.0049521
Stanyon R, Stone G (2008) Phylogenomic analysis by chromosome sorting and painting. Methods Mol Biol 422:13–29. doi:10.1007/978-1-59745-581-7_2
Stanyon R, Consigliere S, Muller S, Morescalchi A, Neusser M, Wienberg J (2000) Fluorescence in situ hybridization (FISH) maps chromosomal homologies between the dusky titi and squirrel monkey. Am J Primatol 50:95–107
Ventura M, Weigl S, Carbone L, Cardone MF, Misceo D, Teti M, D’Addabbo P, Wandall A, Bjorck E, de Jong PJ, She X, Eichler EE, Archidiacono N, Rocchi M (2004) Recurrent sites for new centromere seeding. Genome Res 14:1696–1703
Ventura M, Antonacci F, Cardone MF, Stanyon R, D’Addabbo P, Cellamare A, Sprague LJ, Eichler EE, Archidiacono N, Rocchi M (2007) Evolutionary formation of new centromeres in macaque. Science 316:243–246
Yonenaga-Yassuda Y, Chu TH (1985) Chromosome banding patterns of Saimiri vanzolinii Ayres. Papiéis Avulsos de Zoologia, Museu de Zoologia da Universidade de São Paulo 36:165–168
Acknowledgements
The authors would like to thank the following persons for their assistance in obtaining and processing of specimens used in this study: Larisa S. Biltueva, Institute of Molecular and Cellular Biology, Novosibirsk; Camila do Nascimento Moreira and Yatiyo Yonenaga-Yassuda, Departamento de Genética e Biologia Evolutiva, Instituto de Biociências, Universidade de São Paulo; June Bellizzi (Assistant Director Zoological Affairs at Catoctin Zoo and Wildlife Preserve), and Richard Hahn (Director, Catoctin Zoo and Wildlife Preserve). This work was supported by PRIN (Progetti di Interesse Nazionale) to RS and by a grant from “Conselho Nacional de Desenvolvimento Cientifico e Tecnològico” (CNPq) to MS (process 407262/2013-0).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
No experimental protocols called for the handling of any animals because the cell lines used were kindly provided by individuals in other institutions.
Conflict of interest
The authors declare that they have no conflict of interests.
Rights and permissions
About this article
Cite this article
Chiatante, G., Capozzi, O., Svartman, M. et al. Centromere repositioning explains fundamental number variability in the New World monkey genus Saimiri . Chromosoma 126, 519–529 (2017). https://doi.org/10.1007/s00412-016-0619-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-016-0619-0