Abstract
We describe a karyotypic polymorphism on the zebra finch Z chromosome. This polymorphism was discovered because of a difference in the position of the centromere and because it occurs at varying frequencies in domesticated colonies in the USA and Germany and among two zebra finch subspecies. Using DNA fluorescent in situ hybridization to map specific Z genes and measurements of DNA replication, we show that this polymorphism is the result of a large pericentric inversion involving the majority of the chromosome. We sequenced a likely breakpoint for the inversion and found many repetitive sequences. Around the breakpoint, there are numerous repetitive sequences and several copies of PAK3 (p21-activated kinase 3)-related sequences (PAK3Z) which showed testes-specific expression by RT-PCR. Our findings further suggest that the sequenced genome of the zebra finch may be derived from a male heterozygote for the Z chromosome polymorphism. This finding, in combination with regional differences in the frequency of the polymorphism, has important consequences for future studies using zebra finches.
Similar content being viewed by others
Introduction
The zebra finch (Taeniopygia guttata), an oscine songbird (Order: Passeriformes), is an important model organism for research on brain sexual differentiation, learning, neurogenesis, and behavioral neurobiology. Recently, the utility of this model species has been enhanced by the sequencing of the zebra finch genome (Warren et al. 2010), new methods for producing transgenic finches (Agate et al. 2009), and the availability of numerous genome resources (Replogle et al. 2008).
The sex chromosome constitution in birds is ZZ (male)/ZW (female), the inverse of mammals (XY:male, XX:female). Although the gene content of the Z chromosome is similar across bird species, the zebra finch Z chromosome shows evidence of several rearrangements relative to that of chicken (Itoh et al. 2006). Other differences have also been detected in the Z chromosome of these species: (1) the existence of W chromosome repetitive sequence (ZBM)-related sequences on Z chromosome in zebra finch, but not in chicken (Itoh et al. 2008); (2) the Z-linked gene, Fem1c gene, has a homologous sequence on W chromosome in zebra finch, but not in other birds (Itoh et al. 2009a); and (3) the zebra finch does not have the MHM sequence and regional dosage compensation seen in chickens (Itoh et al. 2007, 2010). Also, as a unique feature of zebra finch species, a germline-restricted chromosome exists only in zebra finch, not in chicken (Pigozzi and Solari 1998; Itoh et al. 2009b).
Heteromorphic sex chromosomes are generally thought to have arisen because of the emergence of a dominant sex-determining gene on an autosome (Charlesworth et al. 2005; Graves 2006, 2010). In mammals, the brain gene Sox3 appears to have mutated into a proto-Sry, adopting a testis-determining function. Changes in the proto-Y chromosome near Sry led to divergence in sequence from the X chromosome and loss of recombination around this region. The non-recombining region accumulated transposable elements, repetitive sequences, mutation/deletion, and male advantageous genes. The non-recombining region gradually grew to include most of the Y chromosome. The size of the chromosome then decreased because of the accumulation of mutations, loss of gene function, and elimination of sequences. Comparable changes are theorized to have occurred in the avian lineage, except that a dominant female-determining mutation is thought to have occurred on the proto-W chromosome.
Karyotypic polymorphisms (translocations, inversions, duplications, and deletions) are common in nature. These chromosome rearrangements sometimes have a large phenotypic effect and tend to be eliminated from the population. For example, in humans, a 60-Mb inversion occurring in the X chromosome led to X-linked congenital adrenal hypoplasia with hypogonadotropic hypogonadism in four related males in a pedigree (Skinningsrud et al. 2009). On the other hand, there are also cases of stable karyotypic polymorphisms (Schorderet et al. 1991). In some cases, these polymorphisms have no known phenotypic effect. For example, zebra finch chromosome 6 has two forms (telocentric and submetacentric) that exist in domesticated populations in all possible combinations (telo/telo, telo/sub, sub/sub; Itoh and Arnold 2005). Another interesting example is the white-throated sparrow (Zonotrichia albicollis) which has an inversion polymorphism of chromosome 2 that is correlated with variation in plumage and social behavior (Thomas et al. 2008). Karytopic polymorphisms have also been suggested to play an important role in the speciation process (Price 2008; Kirkpatrick and Barton 2006). Recombination can be suppressed within the inversion (as observed in the white-throated sparrow; Thomas et al. 2008), leading to an accumulation of differences that can cause genomic incompatibilities among individuals with different chromosomal types.
In this study, we have identified a karyotypic polymorphism in zebra finch Z chromosome using metaphase spreads and fluorescent in situ hybridization (FISH) mapping. Two types of Z chromosomes were found to be widely distributed in both domesticated and wild zebra finch populations and have accumulated repetitive sequences and a male advantageous gene around the inversion breakpoint. The insertion of repetitive sequences and accumulation of male-advantageous genes reflect changes that are predicted by theory to occur in evolving sex chromosomes.
Materials and methods
FISH and DNA replication assay
Metaphase chromosome sets were prepared from day 5–8 embryos or skin samples from adults according to Itoh and Arnold (2005). FISH was carried out using the methods described in two previous papers (Itoh and Arnold 2005, 2006). The bacterial artificial chromosome (BAC) probes for FISH mapping were listed in Itoh et al. (2006).
For the DNA replication assay, zebra finch embryonic fibroblasts were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% fetal bovine serum and 2% chicken serum at 37°C. When the cells were 30–40% confluent, thymidine (Sigma; final concentration 300 μg/ml) was added to the dish and cells were incubated for an additional 16–18 h. The cells were washed three times in fresh medium and incubated in the fourth change of medium for 4 h. After 5-Bromo 2′-deoxyuridine (BrdU) was added to the cultures at a final concentration of 30 μM, cells were incubated for 60 min. Colcemid (Gibco BRL; final concentration 5 μg/ml) was added to the culture and the plates were incubated for additional 2 h, followed by harvesting cells and preparation of metaphase chromosome sets. The slides were baked at 63°C for 2 h, the DNA denatured in 70% formamide-2xSSC at 72°C for 2 min, and immediately chilled in ice-cold 70% ethanol for 3 min. After dehydrating the slides with a series of ethanols (70%, 90%, and 100%) for 3 min each, FISH hybridization was carried out using the methods described previously (Itoh and Arnold 2005; Itoh et al. 2006). The BrdU-incorporated DNA was detected with mouse anti-BrdU (first antibody) and anti-mouse Texas Red (second antibody) in the same cells in which FISH signals were detected. DNA was counterstained with DAPI and covered with VECTASHIELD mounting medium (Vector Labs). The timing of BrdU incorporation in this protocol results in labeling of the late-replicating W chromosome as well as late-replicating regions of the Z chromosome.
Genomic PCR for detection of the type of Z chromosome
In order to examine the distribution of the Z inversion among captive and natural zebra finch populations, we developed a PCR assay. Using PCR to predict Z chromosome type, we examined the proportions of Z chromosome types in the samples of three colonies of domesticated birds (UCLA, Rockefeller University, Free University Berlin) as well as in samples of DNA from wild birds in Australia and Lombok/West Timor. Sampling of wild birds is described in Balakrishnan and Edwards (2009). The samples from Lombok/West Timor are from the Timor zebra finch subspecies (T. guttata guttata). Two zebra finch subspecies appear to have diverged around 1 Mya (Balakrishnan and Edwards 2009), but are known to be able to breed with Australian zebra finches (Clayton 1990).
PCR was carried out in a 25 μl mixture containing 0.2 mM each of dNTPs, 2 mM MgCl2, 0.4 μM each of primers, 10 ng genomic DNA, 0.25 μl of Taq polymerase (Invitrogen), and one tenth volume of 10× Taq buffer (Invitrogen). The sequences of primers were ZINVG-F1 (5′-GCA CTG TCC TCT GGG TCT TC-3′) and ZINVG-R1 (5′-ACT CTG GGC TAG AGG CTC CC-3′). The PCR reaction started at 95°C for 4 min before the cycling reaction of 40 cycles of 95°C for 45 s/60°C for 30 s/72°C for 3 min, and then followed by a single reaction at 72°C for 7 min. Ten microliters of the PCR reaction mixture was separated by 1.5% agarose gel electrophoresis in 1× TAE at 80 V.
BAC sequencing, gene prediction, and RT-PCR
Zebra finch BAC clone, 033L23, was isolated from a BAC library made by the Arizona Genomics Institute (www.genome.arizona.edu; Luo et al. 2006). The sequence of 033L23 BAC DNA was determined by the Washington University Genome Center and is now in Genbank under accession no. AC216736 (135,150 bp). Since this BAC was sequenced as four unordered pieces, we arranged the order of working contigs by comparing it to the zebra finch genome (Ensembl: www.ensembl.org) and confirming the order by performing PCR across the BAC fragment ends and sequencing the products. Gene prediction analysis was performed on the BAC sequence by GENSCAN (http://genes.mit.edu/GENSCAN.html).
RT-PCR was carried out in a 25 μl mixture containing 0.2 mM each of dNTPs, 1.5 mM MgCl2, 0.4 μM each of primers, 10 ng genomic DNA, 0.25 μl of Taq polymerase (Invitrogen), and one tenth volume of 10× Taq buffer (Invitrogen) using primers for the PAK3Z gene: PAK3-2F (5′-CTC TGA GCC TCC TCT GGA TG-3′), PAK3-1R (5′-CAG CAG CAC AGA AAG TCT CG-3′). The PCR reaction started at 95°C for 4 min before the cycling reaction of 40 cycles of 95°C for 45 s/60°C for 30 s/72°C for 3 min, and then followed by a single reaction at 72°C for 7 min. Ten microliters of PCR reaction mixture was separated by 1.5% agarose gel electrophoresis in 1× TAE at 80 V.
Results
Existence of two types of Z chromosomes in zebra finch populations
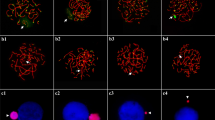
In the process of karyotyping many individuals in our domesticated zebra finch colony, we discovered that there were two morphological types of Z chromosomes: submetacentric Z chromosomes (Zsm) and metacentric Z chromosomes (Zm; Figs. 1 and 2). Although the position of centromere was close in Zsm (39.66% from p-terminus) and Zm (45.12%), these two types of Z chromosomes were clearly distinguishable in DAPI- or Giemsa-stained metaphases (Fig. 2 and Table 1). We previously reported the centromere position of the Z chromosome as 42.6 ± 1.1%, which is approximately an average of the two Z centromere positions (Itoh et al. 2006), probably because we measured and averaged both Zsm and Zm. All three possible pairs of Z chromosomes existed in males: Zsm/Zsm (Fig. 1a), Zm/Zm (Fig. 1b), and Zsm/Zm (Fig. 1c). Based on breeding records, it is likely that Zsm/Zm males can breed even though they have a heteromorphic Z chromosome pair. In females, we found both Zsm and Zm in our colony.
To further analyze the positional difference of the centromere, we mapped several genes along the Z chromosome using FISH. DMRT1 (doublesex and mab-3 related transcription factor 1) and HINT1 (histidine triad nucleotide binding protein 1) were found in different positions (Fig. 3). Whereas the location of DMRT1-encoding BAC was on long arm in both Zsm and Zm, HINT1-encoding BAC localized to the short arm in Zsm and both the short and long arms in Zm chromosome (Fig. 3). The single location of HINT1-encoding BAC on Zsm matches the single location of the HINTZ gene in our previous FISH-based mapping of the zebra finch Z chromosome (Itoh et al. 2006). Thus, the HINT1-encoding BAC 033L23 is from Zsm, and our Z map previously published reflects the gene order of Zsm (Itoh et al. 2006). On the other hand, Zm appeared to contain some arrangement such as an inversion around the HINT1 gene. The unequal signal of BAC 033L23 encoding HINT1 was weaker on the short arm than on the long arm of Zm, suggesting that the majority of the FISH probe sequence localizes to the long arm (Fig. 3). We also examined the location of the late replication site in Zsm and Zm using BrdU incorporation assay. The location of late replicated DNA was obviously different between Zsm and Zm: The signal was on the terminus of long arm in Zsm and on the terminus of short arm in Zm (Fig. 3). By mapping the location of five Z chromosome genes and the late replication locus, we found that the terminal region of short and long arms were switched between Zsm and Zm, leaving the locations of SPIN (spindlin), GHR (growth hormone receptor), and DMRT1 the same (Fig. 4). These results suggest that the one form of the Z chromosome represents a large pericentric inversion of the other, involving the majority of the chromosome.
Location of DMRT1 (028P16, green), HINT1 (033L23, green), and late replication site (red) on Zsm and Zm chromosomes. The signal of 033L23 was split into two spots on Zm chromosome: stronger signal on long arm and weaker signal on short arm. Thus, the origin of 033L23 BAC clone is the Zsm chromosome since there is only one signal. Counter staining: DAPI
Diagram of relative position of FISH signals and late replication site on Zsm and Zm chromosomes. On the Zm chromosome, the 033L23 BAC signal (HINT1 gene) is larger at the major site and smaller at the minor site. We have previously reported the gene locations for the Zsm chromosome (Itoh et al. 2006)
Candidate region of Z chromosome inversion site
Since the BAC clone for HINT1 gene, 033L23, likely contains the breakpoint of the inversion, we sequenced the entire BAC and aligned it to the published zebra finch genome sequence (Fig. 5; Ensembl: www.ensembl.org). The assembly of these sequences resulted in four unordered contigs of lengths 7,022, 17,885, 21,449, and 88,494 bp. Regions of these contigs mapped (BLAT) to zebra finch chromosome Z, chromosome Z random, and chr Un, underscoring the difficulty in the genomic assembly of this region. Although there was a region where these two sequences were highly homologous (Fig. 5), the region around 0–60,000 bp in 033L23 (mainly in the 7,022- and 21,449-bp fragments from original 033L23) was strikingly different from the corresponding region of the published zebra finch genome sequence. Within this region, some sequences were repeated several times in both 033L23 and the published genome. For example, the 3-kb fragment around the 5,851,928- to 5,855,117-bp (Z chromosome) region of the published zebra finch genome was repeated twice in 033L23 BAC sequence at 23,230–26,407 and 32,322–35,514 bp (Fig. 5). Although it is likely that high repeat content has led to errors in the genomic and BAC assembly, the existence of repeat sequences is characteristic of breakpoint regions (Fig. 5).
Alignment of 033L23 sequence (Zsm) to the corresponding zebra finch genome (Zm; from Ensembl: www.ensembl.org). The region between 0 and 50,000 bp of the 033L23 BAC clone should contain the inversion point since there are many repetitive units as an indicator of rearrangement. Asterisk shows the location of DNA fragment where ZINVG-F1 and ZINVG-R1 primers amplify
In an attempt to find the inversion point within this region (033L23, 0–60,000 bp), we designed several primer sets within the candidate regions and performed PCR to amplify genomic DNA. One primer set correlated perfectly with the type of Z chromosome as determined by karyotype in our colony (six males: Zsm/Zsm, seven males: Zsm/Zm, one male: Zm/Zm, three females: Zsm). This primer set amplified a 1-kb fragment from Zsm and a 3-kb fragment from Zm (Fig. 6). Unfortunately, this primer did not straddle the inversion breakpoint but rather detected the existence of a 2-kb insertion sequence (data not shown) which correlated with the type of Z chromosomes.
We further analyzed birds from the Rockefeller University (New York) colony to evaluate the correlation with karyotype (Table 2). In birds from UCLA and Rockefeller, the PCR result matched the karyotype in all cases except two samples in the Zsm/Zsm male group (Table 2), which were birds with the same parents. Thus, we consider that PCR product is a Z chromosome polymorphism by itself, which is a good (93% accurate) but not perfect predictor of the karyotypic inversion.
Geographic variation in inversion frequencies
Using PCR to predict Z chromosome type, we examined the proportions of Z chromosome types in the samples of three colonies of domesticated birds (UCLA, Rockefeller University, Free University Berlin) as well as in samples of DNA from wild birds in Australia and Lombok/West Timor. The samples from Lombok/West Timor were from the smaller Timor zebra finch subspecies (T. guttata guttata), which are known to be able to breed with the Australian zebra finch (Clayton 1990). Based on our PCR assay, we found statistically significant differences in the frequencies of the two chromosomal morphs between the two subspecies (χ 2 = 45.8, p < 0.001) and between German and American domesticated populations (χ 2 = 4.9, p = 0.0269; Table 3, Fig. 7, and Electronic supplementary material Table 1).
Distribution of Zsm and Zm chromosomes in Australia. Bar graph shows the percentage of each chromosomes (gray: Zsm, black: Zm; Electronic supplementary material Table 1). Map was made based on the web site (http://www.aquarius.geomar.de/). The bar for Lombok/West Timor is from Timor zebra finch (T. guttata guttata) samples
Z-linked PAK3 gene
We sought evidence that genes near the inversion breakpoint might have been destroyed or modified because of the arrangement of DNA. Using Genescan gene prediction software, we found four putative genes (genes 1–4) predicted within the 033L23 BAC sequence (Fig. 8). Predicted genes 1 and 2 had homology to PAK3 (p21-activated kinase 3) gene and gene 3 to HINT1 gene, although the predicted gene sequences seem to contain extra exons not related to PAK3 or HINT1 gene (Fig. 8). Z chromosome PAK3-related sequences (PAK3Z) were also found in three separate regions within 0–40 kb of 033L23 BAC clone, where we predicted the existence of inversion breakpoint (Fig. 5). Thus, the PAK3Z gene appears to be influenced by the inversion and was duplicated around the breakpoint (Fig. 8).
Genescan (http://genes.mit.edu/GENSCAN.html) predicted four putative genes on 033L23 BAC sequence. Genes 1 and 2 have a homology to PAK3 and gene 3 has a homology to HINT1. Regions highly homologous to PAK3 and HINT1 are shown above the line. Vertical bars indicate putative exons identified by Genescan, but some of these may not be related to PAK3 and HINT1. The breakpoint of the Z chromosome inversion is probably somewhere within genes 1 or 2
Figure 9 shows the alignment of partial amino acid sequences of PAK3 (human: BAC81128, chimpanzee: BAC81129, Bornean orangutan: BAC81130, mouse: EDL14733, African clawed frog: AAN52281, chicken: XM_420314, and zebra finch: ENSTGUP00000004564) and PAK3Z genes. In zebra finch, the PAK3 gene found on chromosome 4A is highly conserved in a protein sequence relative to several mammals and birds. The PAK3Z genes, which had lower homology to mammalian PAK3 (Fig. 9), can be considered as extra copies. PAK3Z genes in the region of the inversion breakpoint lack some amino acids of the conserved protein. RT-PCR for brain, lung, and gonad samples showed that expression of PAK3Z is restricted to the testes (Fig. 10).
Partial alignment of PAK3 protein sequences. Predicted genes 1 and 2 (Fig. 8) showed high homology to PAK3 from the other species. Asterisks indicate conserved amino acids in all nine sequences
Discussion
We have discovered the existence of two types of Z chromosomes in zebra finch populations, Zsm and Zm, recognizable by the position of centromere. Using FISH mapping and DNA replication timing assay, we found that a large centromeric portion of Z chromosome, comprising a majority of the chromosome, was inverted during evolution. We have also analyzed the sequence of the BAC clone located around the breakpoint of inversion and described a 2-kb insertion fragment that correlates with the type of Z chromosomes. Using PCR to predict the type of Z chromosome, we have analyzed the distribution of the Z chromosome type in a number of domesticated and wild populations and found that the Zsm chromosome is more frequent overall, but that frequencies vary among populations. Based on gene prediction analysis, sequences related to PAK3 (PAK3Z) were located in this Z chromosome inversion breakpoint region, and PAK3Z expression was found in testes.
Based on available data, the arrangement of Zm to Zsm chromosomes can be explained by one large-scale inversion (Fig. 11). The physical map of the Zm chromosome is more similar to the locations of genes and the heterochromatic region (late replication) of the chicken Z chromosome. In this model, the gene order of Zsm would largely match that of Zm.
Dynamic inversion model of the zebra finch Z chromosome. The physical map of Zm chromosome is more similar to the gene location of chicken Z chromosome. Considering the positions of FISH signals and late replication site on Zsm and Zm, one possible explanation for this karyotypic polymorphism is an inversion of the large middle region of Z chromosome. 033L23 BAC probe contains HINT1 gene and shows two signal locations in Zm (major and minor). The heterochromatic region on the long arm of chicken Z chromosome was reported by Hori et al. (1996). A similar region showed a pattern of late replication based on the BrdU incorporation assay (Itoh et al., unpublished data)
Several scenarios could explain the much higher frequency of Zsm, relative to Zm, in the zebra finch mainland population and the contrasting higher frequency of Zm in the Timor subspecies. One explanation is that the Timor subspecies is the result of a small number of founders that were predominantly Zm. Alternatively, the frequency of the two chromosomes could have been similar originally in the two groups, but became different because of divergent selection or genetic drift. The proximity of the breakpoint to the testis-determining gene DMRT1 (Smith et al. 2009) and the duplication of testis-specific (perhaps male-advantageous) PAK3 sequences around the breakpoint hint at genomic changes that could alter male function. Perhaps the least likely scenario is that the polymorphism evolved in the Timor finch subspecies (T. guttata guttata) and was reintroduced to the ancestral Australian population because of reverse migration from Timor to Australia. Population genetic estimates of gene flow based on these same wild populations suggest negligible gene flow between the Lesser Sunda islands and the Australian Mainland (Balakrishnan and Edwards, 2009). Unfortunately, we were unable to test the reliability of the PCR method to predict Z chromosome type for Timor zebra finch, and thus further information is needed on the Z chromosome karyotypic forms in that subspecies.
Intriguingly, the frequencies of the chromosome morphs vary among captive populations in Germany and the USA that are intensively studied for research in neuroscience and behavior. As in the wild populations, differences among captive populations are due to the details of their history. Zebra finches have been reared in captivity for over 100 years. Using neutral markers, Forstmeier et al. (2007) found relatively low but statistically significant genetic differences among captive zebra finch populations. Our study identifies a population difference that could have functional consequences for studies of the brain and behavior. Given the important role of the zebra finch as a model system, a more thorough understanding of genetic variation among natural and captive populations is desirable.
We identified all possible combinations of the two Z chromosome morphs. A remaining puzzle therefore is how this polymorphism persists without causing problems during meiosis. Because the inversion significantly changed the gene order, meiosis would be required to proceed in an irregular manner in order to avoid arrest at the meiotic checkpoint. In males with a heteromorphic Z chromosome pair (Zsm/Zm), pairing of two Z chromosomes may not involve the entire Z chromosomes and thus might be similar to the restricted pairing of the pseudoautosomal regions of sex chromosomes. The pairing could occur either at ends of Z chromosome or large middle portion of the chromosome, leaving the other non-aligned part(s) unpaired. This restricted pairing and crossing over pattern may have forced a change in the DNA sequence of the unpaired region, especially near the inversion breakpoint. The repetitive sequences and multiple PAK3Z sequences near the breakpoint may be evidence for such rearrangement.
In the zebra finch, PAK3-like genes have radiated dramatically (Warren et al. 2010; Kong et al. 2010), including the PAK3-Z chromosome copy found here at the inversion breakpoint. In chicken, the PAK3 gene is found in only a single copy and is autosomal (Ensembl; Kong et al. 2010). The zebra finch PAK3Z described here therefore represents a duplication to the Z chromosome that occurred after the chicken–zebra finch split. The missing amino acid sequences of the PAK3Z protein near the breakpoint (Fig. 12) could represent changes or loss of PAK3 function. Among the 31 PAK3-like genes described in zebra finch (Kong et al. 2010), at least 11 are found on the Z chromosome (with numerous others not yet mapped to chromosomes). Testis expression of PAK3 genes in zebra finch is particularly interesting because the mammalian ortholog shows restricted expression in the central nervous system. The radiation of PAK3s therefore may be related to the evolution of a function in the testis (Kong et al. 2010). The locus described here from the breakpoint region shows testis-specific expression that may reflect the evolution of a sex-specific function, a characteristic evolutionary pattern of sex chromosomes.
Comparison of zebra finch PAK3 protein sequences to chicken. The protein sequence of zebra finch PAK3 located on chromosome 4 has high homology to that of the chicken (99% identity). Predicted genes 1 and 2 from BAC clone 033L23 (Fig. 8; Zsm) and predicted gene 5 from the published zebra finch genome sequence (Zm; Ensembl: www.ensembl.org) have regions homologous to chicken, but lack several amino acid sequences. The percentage of identities to chicken is shown only for the alignment with the highest score
We have analyzed the karyotypes of the father and siblings of the zebra finch from our colony (UCLA Black 17) whose genome sequence has been published (Warren et al. 2010). The Z chromosome component was Zsm/Zm for its father, Zsm/Zsm for one sibling, and Zsm/Zm for the other sibling. Thus, although the mother died before her karyotype could be analyzed, her Z chromosome should have been Zsm. Thus, the Z chromosome pair of Black 17 would have two possibilities: Zsm/Zsm or Zm/Zsm. If Black 17 was Zm/Zsm, the published genome sequence is based on the two types of sequences around the inversion point, which would complicate the assembly of contigs and leave a region of misalignment. Our findings therefore have important implications for the improvement of the zebra finch genome assembly and for the study of genomics in this important study system.
References
Agate RJ, Scott BB, Haripal B, Lois C, Nottebohm F (2009) Transgenic songbirds offer an opportunity to develop a genetic model for vocal learning. Proc Natl Acad Sci USA 106(42):17963–17967
Balakrishnan CN, Edwards SV (2009) Nucleotide variation, linkage disequilibrium and founder-facilitated speciation in wild populations of the zebra finch (Taeniopygia guttata). Genetics 181(2):645–660
Charlesworth D, Charlesworth B, Marais G (2005) Steps in the evolution of heteromorphic sex chromosomes. Heredity 95(2):118–128
Clayton NS (1990) Assortative mating in zebra finch subspecies, Taeniopygia guttata guttata and T. g. castanotis. Philos Trans Biol Sci 330:351–370
Forstmeier W, Segelbacher G, Mueller JC, Kempenaers B (2007) Genetic variation and differentiation in captive and wild zebra finches (Taeniopygia guttata). Mol Ecol 16(19):4039–4050
Graves JA (2006) Sex chromosome specialization and degeneration in mammals. Cell 124(5):901–914
Graves JA (2010) Review: sex chromosome evolution and the expression of sex-specific genes in the placenta. Placenta 31(Suppl):S27–S32
Hori T, Suzuki Y, Solovei I, Saitoh Y, Hutchison N, Ikeda JE, Macgregor H, Mizuno S (1996) Characterization of DNA sequences constituting the terminal heterochromatin of the chicken Z chromosome. Chromosome Res 4(6):411–426
Itoh Y, Arnold AP (2005) Chromosomal polymorphism and comparative painting analysis in the zebra finch. Chromosome Res 13:47–56
Itoh Y, Kampf K, Arnold AP (2006) Comparison of the chicken and zebra finch C chromosomes shows evolutionary rearrangements. Chromosome Res 14(8):805–815
Itoh Y, Melamed E, Yang X, Kampf K, Wang S, Yehya N, Van Nas A, Replogle K, Band MR, Clayton DF, Schadt EE, Lusis AJ, Arnold AP (2007) Dosage compensation is less effective in birds than in mammals. J Biol 6(1):2
Itoh Y, Kampf K, Arnold AP (2008) Molecular cloning of zebra finch W chromosome repetitive sequences: evolution of the avian W chromosome. Chromosoma 117(2):111–121
Itoh Y, Kampf K, Arnold AP (2009a) Disruption of FEM1C-W gene in zebra finch: evolutionary insights on avian ZW genes. Chromosoma 118(3):323–334
Itoh Y, Kampf K, Pigozzi MI, Arnold AP (2009b) Molecular cloning and characterization of the germline-restricted chromosome sequence in the zebra finch. Chromosoma 118(4):527–536
Itoh Y, Replogle K, Kim YH, Wade J, Clayton DF, Arnold AP (2010) Sex bias and dosage compensation in the zebra finch versus chicken genomes: general and specialized patterns among birds. Genome Res 20(4):512–518
Kirkpatrick M, Barton N (2006) Chromosome inversions, local adaptation and speciation. Genetics 173(1):419–434
Kong L, Lovell PV, Heger A, Mello CV, Ponting CP (2010) Accelerated evolution of PAK3- and PIM1-like kinase gene families in the zebra finch, Taeniopygia guttata. Mol Biol Evol 27(8):1923–1934
Luo M, Yu Y, Kim H, Kudrna D, Itoh Y, Agate RJ, Melamed E, Goicoechea JL, Talag J, Mueller C, Wang W, Currie J, Sisneros NB, Wing RA, Arnold AP (2006) Utilization of a zebra finch BAC library to determine the structure of an avian androgen receptor genomic region. Genomics 87(1):181–190
Pigozzi MI, Solari AJ (1998) Germ cell restriction and regular transmission of an accessory chromosome that mimics a sex body in the zebra finch, Taeniopygia guttata. Chromosome Res 6:105–113
Price T (2008) Speciation in birds. Roberts and Company, Greenwood Village, CO, pp 383–393
Replogle K, Arnold AP, Ball GF, Band M, Bensch S, Brenowitz EA, Dong S, Drnevich J, Ferris M, George JM, Gong G, Hasselquist D, Hernandez AG, Kim R, Lewin HA, Liu L, Lovell PV, Mello CV, Naurin S, Rodriguez-Zas S, Thimmapuram J, Wade J, Clayton DF (2008) The Songbird Neurogenomics (SoNG) Initiative: community-based tools and strategies for study of brain gene function and evolution. BMC Genomics 9:131
Schorderet DF, Friedman C, Disteche CM (1991) Pericentric inversion of the X chromosome: presentation of a case and review of the literature. Ann Génét 34(2):98–103
Skinningsrud B, Husebye ES, Gilfillan GD, Frengen E, Erichsen A, Gervin K, Ormerod E, Egeland T, Undlien DE (2009) X-linked congenital adrenal hypoplasia with hypogonadotropic hypogonadism caused by an inversion disrupting a conserved noncoding element upstream of the NR0B1 (DAX1) gene. J Clin Endocrinol Metab 94(10):4086–4093
Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, Doran TJ, Sinclair AH (2009) The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461(7261):267–271
Thomas JW, Cáceres M, Lowman JJ, Morehouse CB, Short ME, Baldwin EL, Maney DL, Martin CL (2008) The chromosomal polymorphism linked to variation in social behavior in the white-throated sparrow (Zonotrichia albicollis) is a complex rearrangement and suppressor of recombination. Genetics 179(3):1455–1468
Warren WC, Clayton DF, Ellegren H, Arnold AP, Hillier LW, Kunstner A, Searle S, White S, Vilella AJ, Fairley S et al (2010) The genome of the zebra finch: special insights into vocal learning and communication. Nature 464(7289):757–762
Acknowledgments
This work was supported by NIH grant DC00217 to A.P. Arnold and a Yamada Science Foundation grant to Y. Itoh. The BAC sequence was determined by the Washington University Genome Center under the supervision of Dr. Wes Warren. Thanks to Silvia Diaz-Perez for assistance with the replication timing experiments and to Robert Agate, Fernando Nottebohm, Constance Scharff, and Jörg Schreiber for providing samples for analysis.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Table 1
Regional difference of Zsm, Zm chromosome numbers in Australia (DOC 28 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Itoh, Y., Kampf, K., Balakrishnan, C.N. et al. Karyotypic polymorphism of the zebra finch Z chromosome. Chromosoma 120, 255–264 (2011). https://doi.org/10.1007/s00412-010-0308-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-010-0308-3