Abstract
Sex chromosomes in different organisms are studied as model systems for chromatin regulation of transcription and epigenetics. Similar to the female X in mammals, the male X chromosome in Drosophila is involved in the process of dosage compensation. However, in contrast to one of the mammalian female X chromosomes undergoing inactivation, the Drosophila male X is transcriptionally upregulated by approximately twofold. The Drosophila male X is a remarkable example for a specialized, transcriptionally hyperactive chromatin domain that facilitates the study of chromatin regulation in the context of transcription, nuclear architecture, and chromatin remodeling. In addition, the rich phenomenology of dosage compensation in Drosophila provides an opportunity to explore the complexities of gene regulation through epigenetic chromatin configurations, histone modifications, and noncoding RNAs. Male-specific lethal (MSL) factors constitute the MSL complex or dosage compensation complex and are important for transcription regulation of X-linked genes. Recent biochemical studies have identified a number of interesting factors that associate with the MSL complex including components of the nuclear pore complex and exosome subunits. Furthermore, global analysis of MSL complex binding showed that MSL complexes are enriched on genes with preferential binding to 3′ end of genes. Taken together, these findings suggest a role of the MSL complex in transcription elongation, RNA processing, and/or nuclear organization.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Dosage compensation in Drosophila—a fascinating story
Different numbers or types of sex chromosomes bring about the distinction between heterogametic and homogametic individuals. Unequal distribution of chromosomes normally results in dramatic changes in gene dosage that can lead to developmental defects or death. Therefore, in parallel with sex chromosomes, animals have evolved mechanisms that ensure equalization of gene expression levels between males and females. These processes evolved independently several times during evolution, and they are collectively termed dosage compensation. The mechanisms of dosage compensation vary largely between different organisms and involve different molecular factors. However, the common theme among all model systems of dosage compensation, which have been analyzed at the molecular level, is their regulation at the level of chromatin.
One of the prominent dosage compensation model systems, which has been genetically and biochemically studied, is the hyperactive X chromosome in male fruit flies. Moreover, dosage compensation as a general principle was first described in Drosophila melanogaster (Bridges 1925). In contrast to mammals (for a review, see Heard and Disteche 2006), in Drosophila females (XX), both X chromosomes are actively transcribed. Accordingly, Drosophila males (XY) have to increase their transcription from the single X chromosome about twofold to compensate for the different gene dosage (see Fig. 1). Molecular factors that have male-specific lethal (MSL) phenotype have been genetically identified (for reviews, see Akhtar 2003; Gilfillan et al. 2004; and Lucchesi et al. 2005). Most of the factors required for dosage compensation in Drosophila have been found in a series of genetic screens for mutants that exhibit male-specific lethality (Belote and Lucchesi 1980; Fukunaga et al. 1975; Hilfiker et al. 1997).
Dosage compensation in Drosophila. Dosage compensation in Drosophila involves an approximately twofold transcriptional upregulation of the single male X chromosome in comparison to female X chromosomes (a). This is mediated by an RNA/protein MSL complex or dosage compensation complex (DCC) which is composed of at least five proteins: MSL1, MSL2, MSL3, MOF, and MLE, and two noncoding RNAs roX1 and roX2 (b). The MSL complex is responsible for hyperacetylation of histone H4 lysine 16 (H4K16) on the male X chromosome
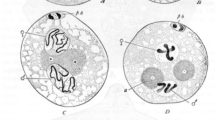
The proteins encoded by the male specific lethal genes are collectively named MSLs and include MSL1, MSL2, MSL3, MLE (maleless), and MOF (m ales absent o n the f irst). Immunofluorescence studies beautifully visualized these proteins colocalizing on hundreds of sites along the male X chromosome, suggesting that they might work together at specific chromosomal sites to promote dosage compensation (see Fig. 2) (Kuroda et al. 1991; Palmer et al. 1993). Indeed, biochemical characterization of MSLs confirmed that they interact physically and form an X-chromosome-specific MSL complex or the dosage compensation complex (DCC) (Copps et al. 1998; Meller et al. 2000; Mendjan et al. 2006; Scott et al. 2000; Smith et al. 2000). Moreover, two noncoding RNAs, roX1 and roX2 (RNA on the X), colocalize specifically with MSLs on the X (Amrein and Axel 1997; Meller et al. 1997). The roX1/roX2 double-mutant flies exhibit an MSL phenotype, and both RNAs were shown to interact with the MSL complex (Franke and Baker 1999; Meller and Rattner 2002; Meller et al. 1997; Smith et al. 2000). MSLs and roX RNAs assemble together at specific nucleation sites on the X chromosome, which have various affinities for the MSL complex (Dahlsveen et al. 2006; Demakova et al. 2003; Kelley et al. 1999).
MSL proteins are enriched on the male X chromosome. Polytene chromosomes from the male third instar larvae were immunostained with antibodies against MSL3 (green) and MSL2 (orange). DNA is visualized by Hoechst (blue). The figure illustrates that MSL proteins specifically recognize and colocalize on hundreds of sites on the male X chromosome
Thus, the MSL complex is a unique ribonucleoprotein complex that specifically recognizes the male X chromosome and mediates a twofold transcriptional upregulation of most X-linked genes irrespective of their inherent differences in expression levels.
Lessons from MSLs—together they are strong
One of the key questions is how does the MSL complex achieve a twofold increase in transcription on the male X chromosome? One way to get some hints about what activities and molecular mechanisms might be associated with the MSL complex is to analyze its components. MSL proteins contain several domains that are generally implicated in chromatin regulation. MSL1 is a PEHE domain containing protein, MSL2 contains a RING finger and a cysteine-rich cluster, and MSL3 contains a chromo-barrel domain and an MRG domain. MLE and MOF contain enzymatic activities that are associated with the MSL complex. MLE is an RNA/DNA helicase also containing a double-stranded RNA binding domain, and MOF is a histone H4 lysine 16 (H4K16) specific histone acetyltransferase (HAT), which in addition to the HAT domain also contains a chromo-barrel domain and a zinc finger (Fig. 3, Table 1, and references therein). Over the years, several studies using in vitro and in vivo approaches have provided a wealth of information about important interaction regions in MSL proteins as well as roX RNAs. A brief summary of the mapping studies is provided in Table 1 (also see Gilfillan et al. 2004; Rea and Akhtar 2006; and Taipale and Akhtar 2005 for reviews). However, despite several in-depth studies, analysis of MSL proteins continues to provide us with new and interesting insights about the complex properties of MSL proteins. Some of the recent advances are discussed below.
One of the best-characterized proteins of the MSL complex is the HAT MOF that specifically acetylates lysine 16 on histone H4 (H4K16Ac) on the X chromosome (Hilfiker et al. 1997). The HAT activity of MOF has been suggested to directly contribute to the stimulation of transcription and therefore has a direct impact on dosage compensation (Akhtar and Becker 2000; Hilfiker et al. 1997). MOF requires to be incorporated into the MSL complex via interactions with MSL3 and MSL1 to exhibit its full enzymatic activity (Morales et al. 2004). As mentioned above besides the HAT domain, MOF contains a chromo-barrel domain similar to MSL3. Chromo-barrel domains have similarity to chromodomains, which have been shown to bind methylated histones, DNA, and RNA (Bernstein et al. 2006; Bouazoune et al. 2002; Flanagan et al. 2005; Min et al. 2003). Chromodomains comprise an essential part of the histone code hypothesis that explains transcriptional regulation through binding and recruitment of specific regulators dependent on specific combinations of histone modifications (Strahl and Allis 2000). Therefore, the question arises if these domains contribute to something like an X chromosomal histone code. The structure of the MOF chromo-barrel domain has revealed that this domain (also present in MSL3) is structurally distinct from the canonical chromodomains found in numerous chromatin-related proteins (Nielsen et al. 2005). MOF chromo-barrel domain is composed of β-barrels, while the canonical chromodomains have an α + β fold. It is interesting to note that this structure resembles the HP1 chromodomain bound to methylated histone H3 lysine 9 (H3K9) (Nielsen et al. 2002), suggesting that the MOF chromo-barrel domain would not be able to accommodate a methylated histone tail. This poses an interesting question as to the function of MOF chromo-barrel domain. The MOF chromo-barrel domain and its adjacent lysine-rich region are required for RNA binding in vitro (Akhtar et al. 2000; Nielsen et al. 2005). It remains to be seen how this region of MOF mechanistically participates in RNA interaction and whether the chromo-barrel domain may also be able to recognize some other histone modifications.
In contrast to MOF, the chromo-barrel domain of MSL3 might be involved in the binding of methylated residues as the aromatic residues required for coordination of methylated lysines are conserved in MSL3. However, mutations in two of the conserved hydrophobic residues in MSL3 chromo-barrel domain do not lead to any visible functional consequences in flies, suggesting that there is no binding in vivo or that additional residues might contribute to the interaction (Buscaino et al. 2006). What residue could be recognized by MSL3 is still a matter of speculation. Histone H3 lysine 36 (H3K36) is a likely candidate because the chromodomain of EAF3, the yeast homologue of MSL3, has recently been shown to bind this modification (Carrozza et al. 2005; Joshi and Struhl 2005; Keogh et al. 2005). However, experimental evidence for such an interaction by MSL3 chromo-barrel domain is still lacking. It would indeed be an interesting possibility if another histone modification in addition to H4K16Ac would contribute to the X-chromosomal epigenetic code.
Deletion analysis of different domains that contribute to the function of MSL3 brought some unexpected insights about the complexity of dosage compensation (Buscaino et al. 2006; Morales et al. 2005). MSL3 contains an MRG domain (MORF4-related gene family) that is found throughout eukaryotes from yeast to humans, with an unknown function (Bertram and Pereira-Smith 2001). This domain is required for proper targeting and integration of MSL3 into the MSL complex on the X chromosome (Morales et al. 2005). However, this is not sufficient to achieve dosage compensation because the chromo-barrel domain is essential for dosage compensation (Buscaino et al. 2006). Hence, the process of dosage compensation contains at least two functionally distinct stages—firstly, recruitment and targeting of the MSL complex to the X chromosome, and secondly, stimulation of X-linked gene expression (Buscaino et al. 2006).
The multifunctionality of MSL3 illustrates some of the complexities that are involved in the mechanism of dosage compensation. While MOF and MSL3 contain domains clearly involved in chromatin regulation, the role of MSL1 and MSL2 in chromatin regulation is more enigmatic. These proteins interact directly through conserved motifs in the N terminus of MSL1 (Copps et al. 1998). Moreover, the N terminus is also required for the binding of MSL1 to the male X chromosome in vivo and for MSL1 self-association in vitro (Li et al. 2005). Potential MSL1 self-association introduces a perspective that favors a structure-mediated function of the MSL complex. This view is further supported by live cell photo-bleaching experiments with GFP-MSL2, which suggest an extraordinary stable association of MSL2 with the X chromosome (Straub et al. 2005). Consequently, the MSL complex might not be a classical chromatin remodeling complex because at least some of its components do not exhibit the high mobility typical for other chromatin remodeling complexes. It remains possible that stable and specific association of MSLs on X-chromosomal genes induces a specific chromosomal configuration, which is particularly permissive to increased and/or regulated transcription.
In summary, the characterization of various domains in MSL proteins suggests a role of the MSL complex at more than one level of chromatin regulation. However, the existence of an X-chromosomal epigenetic code remains a pure speculation because the specific interactions between chromatin modules and corresponding histone modification have not been found yet.
Walking on the X—tracking the footsteps of MSLs
For many years, one of the major questions about dosage compensation in Drosophila has been how MSLs specifically recognize the X and what are the binding sites of the MSL complex on the X chromosome. This is of general interest because there is no simple X-chromosomally specific DNA sequence that could explain the preference of MSLs for the X instead of autosomes. Polytene chromosome spreads have provided a formidable system for visualization of MSL binding on the male X chromosome (Fig. 2). Hundreds of distinct bands bound by the MSL complex can be reproducibly immunostained by specific antibodies against MSL proteins or roX1/2 RNA fluorescence in situ hybridization (Kuroda et al. 1991; Meller et al. 1997). Two of these sites are roX1 and roX2 genomic loci that are bound strongly by the MSL complex. Initially, it was proposed that these are sites of MSL complex assembly, which then further spreads in cis over other binding sites on the X chromosome (Kelley et al. 1999). The characterization of translocations of X fragments to autosomes has led to an alternative model proposing sites on the X chromosome with different affinities for the MSL complex (Fagegaltier and Baker 2004; Oh et al. 2004). The affinity model suggests that there is an X-specific hierarchy of binding affinities for MSL proteins. In addition, there is also a clear hierarchy in complex assembly on the X chromosome that distinguishes MSL1 and MSL2 as essential for further binding of other complex components (Lucchesi et al. 2005). Consequently, specific MSL recruitment to the X chromosome seems to be a complex interplay between different cis- and trans-acting factors. MSL proteins appear to have intrinsic affinity for X-chromosomal sequences, which is tightly balanced with their expression levels and interaction with roX RNAs, as demonstrated by the observations that overexpression of MSL1 and MSL2 complex can partially rescue male lethality in the absence of roX RNAs (Oh et al. 2003; Park et al. 2002).
The identification of MSL binding sites in more detail required sophisticated mapping approaches because polytene chromosomes, although very useful, provide limited resolution at the molecular level. The development of functional genomic approaches like chromatin immunoprecipitation coupled with DNA microarrays (ChIP-chip) finally allowed the assessment of this question on a global scale. Previous ChIP experiments suggested that the complex binds some dosage-compensated genes in the 3′ coding region, but the question remained whether this also had a global significance (Smith et al. 2001). Three recent studies reported the binding pattern of MSL proteins on the X chromosome using various experimental platforms (Alekseyenko et al. 2006; Gilfillan et al. 2006; Legube et al. 2006). These studies, consistent with earlier observations, revealed the preference of the MSL complex to bind to gene coding regions and in particular to the 3′ end of genes (Alekseyenko et al. 2006; Gilfillan et al. 2006). It is interesting to note that studies comparing the binding profiles of MSL1 in embryos and larvae showed that the binding pattern of MSL1 does not dramatically change at different stages of development (Kotlikova et al. 2005; Legube et al. 2006). This is consistent with the additional finding that preferential targets of the MSL binding might be housekeeping/essential genes that are generally expressed throughout fly development (Gilfillan et al. 2006).
Another intriguing question was whether MSL complex binding coincides with RNA polymerase II (RNA-PolII) activity on X-linked genes. Recent observations suggest a weak or no significant correlation between transcription and MSL complex binding, at least, in salivary glands (Kotlikova et al. 2005; Legube et al. 2006). However, correlation of MSL binding with the active state of gene expression in embryos and different Drosophila cell types was indeed observed (Alekseyenko et al. 2006; Gilfillan et al. 2006; Legube et al. 2006). It is therefore likely that tissue-specific differences may influence binding of MSL complex on the X chromosome and that may also in turn be influenced by the expression status of the X-linked genes (Sass et al. 2003). Clearly, the mode of X-chromosomal recognition by the MSL complex is not simple and probably involves multiple steps. Transcription or degenerate DNA elements or subchromosomal context may all contribute to the X-chromosomal specificity of the MSL complex.
MSLs are not alone—the MSL complex meets other nuclear factors
In 1965, Mukherjee and Beermann analyzed RNA synthesis along the male polytene X chromosome by H3 uridine incorporation and observed increased transcription in comparison to female X chromosomes (Mukherjee and Beermann 1965, 1966). This was the first direct evidence suggesting a role of transcription in dosage compensation. However, despite continuous efforts to discover the components and mechanism of dosage compensation, the question is yet unresolved as to how the MSL complex might impact on transcription.
Our knowledge about the various single components of the MSL complex does not explain how the specific twofold transcriptional upregulation is achieved. We know that the upregulation is dependent on the MSL complex, but a direct link to the basal machinery that modulates gene expression is missing. One possibility could be that there is no such direct link. The mechanism of dosage compensation could involve novel functions affecting general chromatin configuration in a unique manner. The other possibility is that the MSL complex contains or interacts with additional general factors that would make the link to established regulatory chromatin transcription systems.
Various levels of chromatin organization are potential regulation points for the mechanism of dosage compensation. There are several reported cases that link the MSL complex to other chromatin-regulating activities/factors. Firstly, there is the JIL1 histone kinase that has been reported to interact with the MSL complex and enriches the X chromosome for H3S10 phosphorylation (Jin et al. 2000). This histone mark has been implicated in chromatin condensation during mitosis and transcription activation, but which of these roles relates to its function in dosage compensation is unclear (Johansen and Johansen 2006). A recent study further suggests a direct involvement of JIL-1 in dosage compensation of genes on the X chromosome (Lerach et al. 2005).
The second possible connection between dosage compensation and general modulators of chromatin state is the chromatin remodeling factor ISWI. In iswi mutant male larvae, the X chromosome appears puffed, and this phenotype can be suppressed by a mutation in mle (Corona et al. 2002). In addition, acetylation at H4K16 by MOF in vitro inhibits binding of ISWI to chromatin templates (Clapier et al. 2001). Accordingly, it has been suggested that there is a balance between ISWI and MSL complex components, which ensures proper chromatin configuration of the X chromosome (Corona et al. 2002). Recent studies suggest a role of the general chromatin factors such as GAGA, Su(var)3–7, and HP-1 in structure maintenance of the X chromosome and in dosage compensation (Greenberg et al. 2004; Spierer et al. 2005).
As discussed above, the MSL complex can be genetically and functionally linked to various general chromatin factors. However, a direct biochemical link of such factors to MSL complex components nor their connection to the mechanism of dosage compensation has not been shown. The recent biochemical purification of the Drosophila and human MSL complexes was therefore bringing some new complementary insights (Mendjan et al. 2006; Smith et al. 2005).
The purification of the Drosophila MSL complexes via tagged MOF and MSL3 proteins leads to a number of unexpected observations. The first surprise was that the purifications did not contain some of the expected suspects like MLE, JIL-1, or ISWI. Weak interactions and/or unfavorable purification conditions may be the reason for their absence. The second surprise was that none of the classical transcription-associated factors was found associated with MOF or MSL3. Instead, there seems to be a core MSL complex (containing MSL1, MSL2, MSL3, and MOF) that copurifies with nuclear pore components or associated proteins (Mtor, Nup153, Nup160, Nup98, and Nup154), interband binding proteins (Z4, Chromator/Chriz), and exosome components (Rrp6, Dis3) as well as a number of novel interaction partners. It is interesting to note that of the several proteins tested in this study, only the depletion of nuclear pore associated proteins Mtor and Nup153 by RNA interference led to the loss of both classical immunostaining of MSL proteins on the X chromosome and dosage compensation of at least a subset of X-linked genes (Mendjan et al. 2006). To date, these proteins are unique, apart from the MSLs themselves, in showing such a severe reduction of MSL staining on the X chromosome as a result of their depletion.
The mechanism of dosage compensation—many hints but no clear answers
Despite much progress in our understanding of MSL interactions with each other or the male X chromosome, it remains a major challenge to decipher the mechanism by which the MSL complex affects gene expression. It has become increasingly clear that dosage compensation involves multiple steps that need to be characterized. The first step comprises the specific recruitment of the MSL complex to the X chromosome. To date, no simple DNA element that would make the X chromosome easily distinguishable from autosomes and allow specific binding of MSLs has been found (Alekseyenko et al. 2006; Dahlsveen et al. 2006; Gilfillan et al. 2006; Legube et al. 2006). This, however, does not mean that the DNA component of chromatin has no role in MSL complex recruitment. Advanced bioinformatic analysis of the X chromosome might lead to the discovery of more complex DNA sequence information that gives some specificity to the X chromosome. In addition, the epigenetic state, specific chromatin configuration, transcription, and degenerated DNA elements could in some combination give the signal for MSL binding.
The second step of dosage compensation is the twofold upregulation of gene expression on the male X (Fig. 4a). Could the histone modification by MOF be the key to the mechanism of dosage compensation? Acetylation of H4K16 by MOF has been shown to cause derepression of transcription either in an in vitro transcription system or when tethered to a reporter gene in yeast; however, the effects were more than twofold in these systems (Akhtar and Becker 2000). There is a possibility that all the other complex components solely help to target and correctly distribute MOF at the X chromosome, while H4K16 acetylation does the actual job. How could this be achieved? The histone code hypothesis predicted correctly, in some cases, that specific effector proteins bind distinct marks (Jenuwein and Allis 2001; Strahl and Allis 2000). However, the receptor (possibly a bromodomain-containing protein) for H4K16Ac has not been found yet. The local distribution of this mark and MSL proteins along gene coding regions implies transcription elongation as a possible regulation point (Alekseyenko et al. 2006; Gilfillan et al. 2006; Legube et al. 2006; Smith et al. 2001). These data are in agreement with the current model of dosage compensation that implies a role of the MSL complex in facilitating transcription elongation (Fig. 4a) (Henikoff and Meneely 1993; Smith et al. 2001). In general, histone acetylation has been already suggested to enhance transcription, but this has not been specifically demonstrated for the H4K16 mark (Orphanides and Reinberg 2000; Svejstrup 2002). It is important to note however that the H4K16 residue is directly implicated in higher-order chromatin structure based on structural studies (Dorigo et al. 2003; Shogren-Knaak et al. 2006; Shogren-Knaak and Peterson 2006). Thus, the effect of H4K16 modification that ultimately leads to dosage compensation may involve levels of chromatin organization and function, which are still poorly understood.
Dosage compensation working models. a The current predominant model of dosage compensation proposes transcription elongation as the main regulating point of the MSL complex. How this is achieved on the molecular level and what is the role of H4K16 acetylation is not yet known (represented by a question mark). H4K16Ac is represented by asterisk, while nucleosomes are represented by gray circles. MSL complex and RNA PolII are represented by purple and orange ovals, respectively. X-chromosomal transcript is represented by gray wiggled line. b MSL proteins are enriched on the 3′ end of X-linked genes, and they have been found to be associated with components of the nuclear pore complex (NPC) (Alekseyenko et al. 2006; Gilfillan et al. 2006; Mendjan et al. 2006; Smith et al. 2001). Consequently, speculative models for the molecular mechanism of dosage compensation can be envisaged that could stimulate our thinking about how MSLs might affect transcription elongation. Model 1 proposes that MSL–NPC interactions might create a specialized X-chromosomal compartment or domain. This model is based on observations in yeast, where the orthologue of Mtor (Mlp1) has been shown to bind transcriptionally active genes (Casolari et al. 2005). Furthermore, transcriptional activity of a gene has the potential to be modulated at the nuclear periphery (Cabal et al. 2006; Taddei et al. 2006). In manner similar to yeast, nuclear pore components in Drosophila may also facilitate modulation of transcriptional activity. This can be achieved by organizing chromatin into functional domains. The X chromosome may therefore require such organization to create functional territories where the MSL complex can act cooperatively to regulate the expression of many X-linked genes. Model 2 proposes that MSLs may link the X chromosome with the nuclear exosome and the NPC and therefore facilitate coupling of transcription elongation and posttranscriptional events such as RNA processing or export. Another interesting possibility is that nuclear pore components like Mtor and/or Nup153 might be involved in efficient processing and maturation of roX RNAs and their efficient incorporation into the MSL complex. The question remains whether these effects are really a consequence of physical tethering of chromatin and/or RNA to the pore or whether dynamic behavior of pore components facilitates such interaction and therefore does not need to occur at the pore. Model 3 therefore suggests a nuclear pore independent function of Mtor and Nup153 in gene regulation. The models 1–3 are not mutually exclusive, and depending on the circumstance, one or more modules may contribute to fine tune gene activity
The local enrichment of MSLs at the 3′ end of genes supports the transcription elongation model (Fig. 4a); however, the mere presence of MSL proteins at the 3′ end of genes is not sufficient to explain mechanistically how MSLs might help facilitate efficient elongation. Integration of the recent data concerning copurification of nuclear pore components and exosome components with MSL proteins into the elongation model of dosage compensation may help to conceptualize how this might be achieved (Fig. 4b).
One model, drawn from analogies to yeast, is that nuclear pore components may influence dosage compensation through chromatin loop organization and spatial positioning (Casolari et al. 2005; Feuerbach et al. 2002). Mtor and Nup153 in Drosophila Schneider cells could affect specific gene or chromosomal domain organization that is required for subsequent mechanisms of chromatin regulation (Fig. 4b, model 1). This model implies that Mtor and Nup153 are indirectly involved in the upregulation of transcription by participating in a higher hierarchical level of chromatin organization and compartmentalization. How important this aspect can be for optimal gene expression was nicely demonstrated in yeast for galactose-inducible genes that require association with the nuclear pore for optimal induction (Cabal et al. 2006; Taddei et al. 2006). It is interesting to note that for HKX1 gene, localization to the nuclear periphery required sequences at the 3′ end of the gene (Taddei et al. 2006). There are some intriguing parallels with enriched binding of MSL complex also at the 3′ end of genes (Alekseyenko et al. 2006; Gilfillan et al. 2006; Smith et al. 2001). However, the significance of this enrichment remains to be explored.
The second model is that Mtor/Nup153 might have a more direct role on gene expression machinery by linking X-chromosomal transcripts and regulatory exosome components (Fig. 4b, model 2). Most of the data concerning the role of the exosome in RNA surveillance, coupling with transcription elongation, and collaboration with Mlp/Mtor have been obtained in yeast (Sommer and Nehrbass 2005). The exosome components Rrp6 and Dis3 participate at different steps of mRNA maturation, and they collaborate with the Mlps at essential steps of gene expression (Galy et al. 2004). Recent work has linked the exosome (Rrp6 subunit) to the TREX complex that is thought to couple transcription with export (Libri et al. 2002; Zenklusen et al. 2002). Moreover, the exosome is recruited to active genes through interactions with transcription elongating factors (Andrulis et al. 2002). The Rrp6 subunit has been implicated in quality control of mRNP particles already at a very early step during transcription (Casolari et al. 2004; Hilleren et al. 2001; Libri et al. 2002). The factors that ultimately link the exosome and Mlp proteins are not known in yeast, and whether the MSLs might fulfill this function in Drosophila is a matter of speculation.
What would be the role of the MSL proteins in this context? At least three MSLs (MOF, MSL3, and MLE) have been shown to bind RNA (Akhtar et al. 2000; Buscaino et al. 2003; Richter et al. 1996). In addition, two of them (MOF and MSL3) copurify Mtor, suggesting that they could have a mediating role between chromatin, transcription, and pre-mRNA processing. There is a possibility that cotranscriptional pre-mRNA processing represents the point of MSL regulation. The MSL complex could specifically facilitate spatial, cotranscriptional coupling between events in gene expression of X-chromosomal genes. The exosome is involved in the processing and quality control of various mRNPs (Hilleren et al. 2001; Zenklusen et al. 2002). Consequently, it might also be required for processing and subsequent assembly of roX RNAs into the MSL complex. roX2 RNA has been shown to undergo variable splicing that contributes to its function in dosage compensation (Park 2005), raising the possibility that other roX RNA processing events might also be critical. It is also interesting to note that a recent study demonstrated the importance of exosome components for Xist RNA processing that is essential for proper X-chromosome inactivation in mammals (Ciaudo et al. 2006). Future studies on roX RNA processing might uncover another interesting parallel between the dosage compensation systems in Drosophila and mammals.
Finally, it can be envisaged that nuclear pore complex members may also function outside of their predominant role as members of the nuclear pore and act as independent submodules for regulating gene activity (Fig. 4b, model 3). Consistent with this hypothesis, a number of nuclear pore components have been shown to display dynamic behavior or exist as soluble pools in the nucleoplasm (Cordes et al. 1997; Griffis et al. 2002, 2004; Hase and Cordes 2003; Hase et al. 2001; Rabut et al. 2004; Zimowska et al. 1997). In this scenario, limited soluble pools of these proteins may bind genic or intergenic regions to function as activators or repressors and in this capacity affect X-chromosomal as well as autosomal gene expression. It is important to note that neither of the above scenarios is mutually exclusive, and combined action of different steps may result in the twofold impact on transcription.
In summary, recent studies have made important advances in our knowledge of dosage compensation in Drosophila. They highlight the complexity behind the twofold upregulation of transcription and various levels of chromosomal regulation and gene expression control that might participate in it. One thing is clear: the dosage compensation process will continue to fascinate us for quite sometime to come.
References
Akhtar A (2003) Dosage compensation: an intertwined world of RNA and chromatin remodelling. Curr Opin Genet Dev 13:161–169
Akhtar A, Becker PB (2000) Activation of transcription through histone H4 acetylation by MOF, an acetyl transferase essential for dosage compensation in Drosophila. Mol Cell 5:367–375
Akhtar A, Becker PB (2001) The histone H4 acetyltransferase MOF uses a C2HC zinc finger for substrate recognition. EMBO Rep 2:113–118
Akhtar A, Zink D, Becker PB (2000) Chromodomains as RNA interaction modules. Nature 407:405–409
Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI (2006) High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev 20:848–857
Amrein H, Axel R (1997) Genes expressed in neurons of adult male Drosophila. Cell 88:459–469
Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT (2002) The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420:837–841
Bashaw GJ, Baker BS (1997) The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell 89:789–798
Belote JM, Lucchesi JC (1980) Control of X chromosome transcription by the maleless gene in Drosophila. Nature 285:573–575
Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD (2006) Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol 26:2560–2569
Bertram MJ, Pereira-Smith OM (2001) Conservation of the MORF4 related gene family: identification of a new chromo domain subfamily and novel protein motif. Gene 266:111–121
Bouazoune K, Mitterweger A, Langst G, Imhof A, Akhtar A, Becker PB, Brehm A (2002) The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. EMBO J 21:2430–2440
Bridges CB (1925) Sex in relation to chromosomes and genes. Am Nat 56:51–63
Buscaino A, Kocher T, Kind JH, Holz H, Taipale M, Wagner K, Wilm M, Akhtar A (2003) MOF-regulated acetylation of MSL-3 in the Drosophila dosage compensation complex. Mol Cell 11:1265–1277
Buscaino A, Legube G, Akhtar A (2006) X-chromosome targeting and dosage compensation are mediated by distinct domains in MSL-3. EMBO Rep 7:531–538
Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, Nehrbass U (2006) SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441:770–773
Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581–592
Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA (2004) Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117:427–439
Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA (2005) Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev 19:1188–1198
Chang KA, Kuroda MI (1998) Modulation of MSL1 abundance in female Drosophila contributes to the sex specificity of dosage compensation. Genetics 150:699–709
Ciaudo C, Bourdet A, Cohen-Tannoudji M, Dietz HC, Rougeulle C, Avner P (2006) Nuclear mRNA degradation pathway(s) are implicated in Xist regulation and X chromosome inactivation. PLoS Genet 2:e94
Clapier CR, Langst G, Corona DF, Becker PB, Nightingale KP (2001) Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol Cell Biol 21:875–883
Copps K, Richman R, Lyman LM, Chang KA, Rampersad-Ammons J, Kuroda MI (1998) Complex formation by the Drosophila MSL proteins: role of the MSL2 RING finger in protein complex assembly. EMBO J 17:5409–5417
Cordes VC, Reidenbach S, Rackwitz HR, Franke WW (1997) Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J Cell Biol 136:515–529
Corona DF, Clapier CR, Becker PB, Tamkun JW (2002) Modulation of ISWI function by site-specific histone acetylation. EMBO Rep 3:242–247
Dahlsveen IK, Gilfillan GD, Shelest VI, Lamm R, Becker PB (2006) Targeting determinants of dosage compensation in Drosophila. PLoS Genet 2:e5
Demakova OV, Kotlikova IV, Gordadze PR, Alekseyenko AA, Kuroda MI, Zhimulev IF (2003) The MSL complex levels are critical for its correct targeting to the chromosomes in Drosophila melanogaster. Chromosoma 112:103–115
Dorigo B, Schalch T, Bystricky K, Richmond TJ (2003) Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J Mol Biol 327:85–96
Fagegaltier D, Baker BS (2004) X chromosome sites autonomously recruit the dosage compensation complex in Drosophila males. PLoS Biol 2:e341
Feuerbach F, Galy V, Trelles-Sticken E, Fromont-Racine M, Jacquier A, Gilson E, Olivo-Marin JC, Scherthan H, Nehrbass U (2002) Nuclear architecture and spatial positioning help establish transcriptional states of telomeres in yeast. Nat Cell Biol 4:214–221
Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S (2005) Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 438:1181–1185
Franke A, Baker BS (1999) The rox1 and rox2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol Cell 4:117–122
Fukunaga A, Tanaka A, Oishi K (1975) Maleless, a recessive autosomal mutant of Drosophila melanogaster that specifically kills male zygotes. Genetics 81:135–141
Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U (2004) Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116:63–73
Gebauer F, Grskovic M, Hentze MW (2003) Drosophila sex-lethal inhibits the stable association of the 40S ribosomal subunit with msl-2 mRNA. Mol Cell 11:1397–1404
Gilfillan GD, Dahlsveen IK, Becker PB (2004) Lifting a chromosome: dosage compensation in Drosophila melanogaster. FEBS Lett 567:8–14
Gilfillan GD, Straub T, de Wit E, Greil F, Lamm R, van Steensel B, Becker PB (2006) Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev 20:858–870
Greenberg AJ, Yanowitz JL, Schedl P (2004) The Drosophila GAGA factor is required for dosage compensation in males and for the formation of the male-specific-lethal complex chromatin entry site at 12DE. Genetics 166:279–289
Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA (2002) Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol Biol Cell 13:1282–1297
Griffis ER, Craige B, Dimaano C, Ullman KS, Powers MA (2004) Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription-dependent mobility. Mol Biol Cell 15:1991–2002
Gu W, Wei X, Pannuti A, Lucchesi JC (2000) Targeting the chromatin-remodeling MSL complex of Drosophila to its sites of action on the X chromosome requires both acetyl transferase and ATPase activities. EMBO J 19:5202–5211
Hase ME, Cordes VC (2003) Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol Biol Cell 14:1923–1940
Hase ME, Kuznetsov NV, Cordes VC (2001) Amino acid substitutions of coiled-coil protein Tpr abrogate anchorage to the nuclear pore complex but not parallel, in-register homodimerization. Mol Biol Cell 12:2433–2452
Heard E, Disteche CM (2006) Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev 20:1848–1867
Henikoff S, Meneely PM (1993) Unwinding dosage compensation. Cell 72:1–2
Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC (1997) mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J 16:2054–2060
Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH (2001) Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413:538–542
Jenuwein T, Allis CD (2001) Translating the histone code. Science 293:1074–1080
Jin Y, Wang Y, Johansen J, Johansen KM (2000) JIL-1, a chromosomal kinase implicated in regulation of chromatin structure, associated with the male specific lethal (MSL) dosage compensation complex. J Cell Biol 149:1005–1010
Johansen KM, Johansen J (2006) Regulation of chromatin structure by histone H3S10 phosphorylation. Chromosome Res 14:393–404
Joshi AA, Struhl K (2005) Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell 20:971–978
Kelley RL, Solovyeva I, Lyman LM, Richman R, Solovyev V, Kuroda MI (1995) Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell 81:867–877
Kelley RL, Wang J, Bell L, Kuroda MI (1997) Sex lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature 387:195–199
Kelley RL, Meller VH, Gordadze PR, Roman G, Davis RL, Kuroda MI (1999) Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 98:513–522
Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, Weissman JS, Hughes TR, Strahl BD, Grunstein M, Greenblatt JF, Buratowski S, Krogan NJ (2005) Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123:593–605
Kotlikova IV, Demakova OV, Semeshin VF, Shloma VV, Boldyreva LV, Kuroda MI, Zhimulev IF (2005) The Drosophila dosage compensation complex binds to polytene chromosomes independently of developmental changes in transcription. Genetics 972:963–974
Kuroda MI, Kernan MJ, Kreber R, Ganetzky B, Baker BS (1991) The maleless protein associates with the X chromosome to regulate dosage compensation in Drosophila. Cell 66:935–947
Lee CG, Chang KA, Kuroda MI, Hurwitz J (1997) The NTPase/helicase activities of Drosophila maleless, an essential factor in dosage compensation. EMBO J 16:2671–2681
Legube G, McWeeney SK, Lercher MJ, Akhtar A (2006) X-chromosome-wide profiling of MSL-1 distribution and dosage compensation in Drosophila. Genes Dev 20:871–883
Lerach S, Zhang W, Deng H, Bao X, Girton J, Johansen J, Johansen KM (2005) JIL-1 kinase, a member of the male-specific lethal (MSL) complex, is necessary for proper dosage compensation of eye pigmentation in Drosophila. Genesis 43:213–215
Li F, Parry DA, Scott MJ (2005) The amino-terminal region of Drosophila MSL1 contains basic, glycine-rich, and leucine zipper-like motifs that promote X chromosome binding, self-association, and MSL2 binding, respectively. Mol Cell Biol 25:8913–8924
Libri D, Dower K, Boulay J, Thomsen R, Rosbash M, Jensen TH (2002) Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol Cell Biol 22:8254–8266
Lucchesi JC, Kelly WG, Panning B (2005) Chromatin remodeling in dosage compensation. Annu Rev Genet 39:615–651
Lyman LM, Copps K, Rastelli L, Kelley RL, Kuroda MI (1997) Drosophila male-specific lethal-2 protein: structure/function analysis and dependence on MSL-1 for chromosome association. Genetics 147:1743–1753
Meller V, Rattner BP (2002) The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J 21:1084–1091
Meller VH, Wu KH, Roman G, Kuroda MI, Davis RL (1997) roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell 88:445–457
Meller VH, Gordadze PR, Park Y, Chu X, Stuckenholz C, Kelley RL, Kuroda MI (2000) Ordered assembly of roX RNAs into MSL complexes on the dosage-compensated X chromosome in Drosophila [in process citation]. Curr Biol 10:136–143
Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermuelen M, Buscaino A, Duncan K, Mueller J, Wilm M, Stunnenberg HG, Saumweber H, Akhtar A (2006) Nuclear pore components are involved in the transcriptional regulation of dosage compensation complex in Drosophila. Molecular Cell 21:1–13
Min J, Zhang Y, Xu RM (2003) Structural basis for specific binding of polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev 17:1823–1828
Morales V, Straub T, Neumann MF, Mengus G, Akhtar A, Becker PB (2004) Functional integration of the histone acetyltransferase MOF into the dosage compensation complex. EMBO J 23:2258–2268
Morales V, Regnard C, Izzo A, Vetter I, Becker PB (2005) The MRG domain mediates the functional integration of MSL3 into the dosage compensation complex. Mol Cell Biol 25:5947–5954
Mukherjee AS, Beermann W (1965) Synthesis of RNA by the X-chromosomes of Drosophila melanogaster and the problem of dosage compensation. Nature 207:785–786
Mukherjee AS, Beermann W (1966) Dosage compensation in Drosophila: an autoradiographic study. The Nucleus 9:83–96
Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED (2002) Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416:103–107
Nielsen PR, Nietlispach D, Buscaino A, Warner RJ, Akhtar A, Murzin AG, Murzina NV, Laue ED (2005) Structure of the chromo barrel domain from the MOF acetyltransferase. J Biol Chem 280:32326–32331
Oh H, Park Y, Kuroda MI (2003) Local spreading of MSL complexes from roX genes on the Drosophila X chromosome. Genes Dev 17:1334–1339
Oh H, Bone JR, Kuroda MI (2004) Multiple classes of MSL binding sites target dosage compensation to the X chromosome of Drosophila. Curr Biol 14:481–487
Orphanides G, Reinberg D (2000) RNA polymerase II elongation through chromatin. Nature 407:471–475
Palmer MJ, Mergner VA, Richman R, Manning JE, Kuroda MI, Lucchesi JC (1993) The male-specific lethal-one (msl-1) gene of Drosophila melanogaster encodes a novel protein that associates with the X chromosome in males. Genetics 134:545–557
Park Y, Kelley RL, Oh H, Kuroda MI, Meller VH (2002) Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science 298:1620–1623
Park Y, Oh H, Meller V, Kuroda MI (2005) Variable splicing of non-coding roX2 RNAs influences targeting of MSL dosage compensation complexes in Drosophila. RNA Biology 2:157–164
Rabut G, Doye V, Ellenberg J (2004) Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol 6:1114–1121
Rea S, Akhtar A (2006) MSL proteins and the regulation of gene expression. Curr Top Microbiol Immunol 310:117–140
Reenan R, Hanrahan CJ, Barry G (2000) The mle(napts) RNA helicase mutation in Drosophila results in a splicing catastrophe of the para Na+ channel transcript in a region of RNA editing. Neuron 25:139–149
Richter L, Bone JR, Kuroda MI (1996) RNA-dependent association of the Drosophila maleless protein with the male X chromosome. Genes Cells 1:325–336
Sass GL, Pannuti A, Lucchesi JC (2003) Male-specific lethal complex of Drosophila targets activated regions of the X chromosome for chromatin remodeling. Proc Natl Acad Sci USA 100:8287–8291
Scott MJ, Pan LL, Cleland SB, Knox AL, Heinrich J (2000) MSL1 plays a central role in assembly of the MSL complex, essential for dosage compensation in Drosophila. EMBO J 19:144–155
Shogren-Knaak M, Peterson CL (2006) Switching on chromatin: mechanistic role of histone h4-k16 acetylation. Cell Cycle 5:1361–1365
Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL (2006) Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311:844–847
Smith ER, Pannuti A, Gu W, Steurnagel A, Cook RG, Allis CD, Lucchesi JC (2000) The drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation [in process citation]. Mol Cell Biol 20:312–318
Smith ER, Allis CD, Lucchesi JC (2001) Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J Biol Chem 276:31483s–31486s
Smith ER, Cayrou C, Huang R, Lane WS, Cote J, Lucchesi JC (2005) A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol 25:9175–9188
Sommer P, Nehrbass U (2005) Quality control of messenger ribonucleoprotein particles in the nucleus and at the pore. Curr Opin Cell Biol 17:294–301
Spierer A, Seum C, Delattre M, Spierer P (2005) Loss of the modifiers of variegation Su(var)3–7 or HP1 impacts male X polytene chromosome morphology and dosage compensation. J Cell Sci 118:5047–5057
Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403:41–45
Straub T, Neumann MF, Prestel M, Kremmer E, Kaether C, Haass C, Becker PB (2005) Stable chromosomal association of MSL2 defines a dosage-compensated nuclear compartment. Chromosoma 114:352–364
Stuckenholz C, Meller VH, Kuroda MI (2003) Functional redundancy within roX1, a noncoding RNA involved in dosage compensation in Drosophila melanogaster. Genetics 164:1003–1014
Svejstrup JQ (2002) Chromatin elongation factors. Curr Opin Genet Dev 12:156–161
Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM (2006) Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441:774–778
Taipale M, Akhtar A (2005) Chromatin mechanisms in Drosophila dosage compensation. Prog Mol Subcell Biol 38:123–149
Zenklusen D, Vinciguerra P, Wyss JC, Stutz F (2002) Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol Cell Biol 22:8241–8253
Zimowska G, Aris JP, Paddy MR (1997) A Drosophila Tpr protein homolog is localized both in the extrachromosomal channel network and to nuclear pore complexes. J Cell Sci 110(Pt 8):927–944
Acknowledgements
We are very grateful to Max Scott, Wolfram Antonin, and Kent Duncan for their critical reading of this manuscript. We thank the members of the lab for their helpful discussions and Maj Britt Hansen for the help with the figures. We also thank the referees for their helpful suggestions. This work was supported by the “Epigenome” NoE under EU-FP6 and DFG “Epigenetics”.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by T. Orr-Weaver
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Mendjan, S., Akhtar, A. The right dose for every sex. Chromosoma 116, 95–106 (2007). https://doi.org/10.1007/s00412-006-0089-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-006-0089-x