Abstract
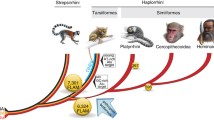
Long interspersed nuclear elements (LINEs) comprise about 21% of the human genome (of which L1 is most abundant) and are preferentially accumulated in AT-rich regions, as well as the X and Y chromosomes. Most knowledge of L1 distribution in mammals is restricted to human and mouse. Here we report the first investigation of L1 distribution in the genomes of a wide variety of eutherian mammals, including species in the two basal clades, Afrotheria and Xenarthra. Our results show L1 accumulation on the X of all eutherian mammals, an observation consistent with an ancestral involvement of these elements in the X-inactivation process (the Lyon repeat hypothesis). Surprisingly, conspicuous accumulation of L1 in AT-rich regions of the genome was not observed in any species outside of Euarchontoglires (represented by human, mouse and rabbit). Although several features were common to most species investigated, our comprehensive survey shows that the patterns observed in human and mouse are, in many aspects, far from typical for all mammals. We discuss these findings with reference to models that have previously been proposed to explain the AT distribution bias of L1 in human and mouse, and how this relates to the evolution of these elements in other eutherian genomes.

Similar content being viewed by others
References
Boissinot S, Furano AV (2001) Adaptive evolution in LINE-1 retrotransposons. Mol Biol Evol 18:2186–2194
Boissinot S, Chevret P, Furano AV (2000) L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol 17:915–928
Boissinot S, Entezam A, Furano AV (2001) Selection against deleterious LINE-1-containing loci in the human lineage. Mol Biol Evol 18:926–935
Boumil RM, Lee JT (2001) Forty years of decoding the silence in X-chromosome inactivation. Hum Mol Genet 10:2225–2232
Boyle AL, Ballard SG, Ward DC (1990) Differential distribution of long and short interspersed element sequences in the mouse genome: chromosome karyotyping by fluorescence in situ hybridization. Proc Natl Acad Sci USA 87:7757–7761
Britten RJ (1996) DNA sequence insertion and evolutionary variation in gene regulation. Proc Natl Acad Sci USA 93:9374–9377
Burton FH, Loeb DD, Voliva CF, Martin SL, Edgell MH, Hutchison CA III (1986) Conservation throughout mammalia and extensive protein-encoding capacity of the highly repeated DNA long interspersed sequence one. J Mol Biol 187:291–304
Caceres M, Puig M, Ruiz A (2001) Molecular characterization of two natural hotspots in the Drosophila buzzatii genome induced by transposon insertions. Genome Res 11:1353–1364
Delsuc F, Scally M, Madsen O, Stanhope MJ, de Jong WW, Catzeflis FM, Springer MS, Douzery EJ (2002) Molecular phylogeny of living xenarthrans and the impact of character and taxon sampling on the placental tree rooting. Mol Biol Evol 19:1656–1671
Dewannieux M, Esnault C, Heidmann T (2003) LINE-mediated retrotransposition of marked Alu sequences. Nat Genet 35:41–48
Dimitri P, Junakovic N (1999) Revising the selfish DNA hypothesis: new evidence on accumulation of transposable elements in heterochromatin. Trends Genet 15:123–124
Dobigny G (2002) Spéciation chromosomique chez les espèces ouest-africaines de Taterillus (Rodentia, Gerbillinae): implications systématiques et biogéographiques, hypotheses génomiques. PhD Thesis, Muséum National d’Histoire Naturelle, Paris, France
Dorner M, Paabo S (1995) Nucleotide sequence of a marsupial LINE-1 element and the evolution of placental mammals. Mol Biol Evol 12:944–948
Duvernell DD, Turner BJ (1998) Swimmer 1, a new low-copy-number LINE family in teleost genomes with sequence similarity to mammalian L1. Mol Biol Evol 15:1791–1793
Evgen’ev MB, Zelentsova H, Poluectova H, Lyozin GT, Veleikodvorskaja V, Pyatkov KI, Zhivotovsky LA, Kidwell MG (2000) Mobile elements and chromosomal evolution in the virilis group of Drosophila. Proc Natl Acad Sci USA 97:11337–11342
Gartler SM, Riggs AD (1983) Mammalian X-chromosome inactivation. Annu Rev Genet 17:155–190
Grinberg MA, Sullivan MM, Benirschke K (1966) Investigation with tritiated thymidine of the relationship between the sex chromosomes, sex chromatin, and the drumstick in the cells of the female nine-banded armadillo, Dasypus novemcinctus. Cytogenetics 5:64–74
Hansen RS (2003) X inactivation-specific methylation of LINE-1 elements by DNMT3B: implications for the Lyon repeat hypothesis. Hum Mol Genet 12:2559–2567
Henikoff S, Malik HS (2002) Centromeres: selfish drivers. Nature 417:227
Kazazian HH Jr (1998) Mobile elements and disease. Curr Opin Genet Dev 8:343–350
Kazazian HH Jr, Goodier JL (2002) LINE drive: retrotransposition and genome instability. Cell 110:277–280
Kidwell MG (2002) Transposable elements and the evolution of genome size in eukaryotes. Genetica 115:49–63
Korenberg JR, Rykowski MC (1988) Human genome organization: Alu, lines, and the molecular structure of metaphase chromosome bands. Cell 53:391–400
van de Lagemaat LN, Landry JR, Mager DL, Medstrand P (2003) Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet 19:530–536
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, Szustakowki J, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921
Levis RW, Ganesan R, Houtchens K, Tolar LA, Sheen FM (1993) Transposons in place of telomeric repeats at a Drosophila telomere. Cell 75:1083–1093
Lorenc A, Makalowski W (2003) Transposable elements and vertebrate protein diversity. Genetica 118:183–191
Lyon MF (1998) X-chromosome inactivation: a repeat hypothesis. Cytogenet Cell Genet 80:133–137
Lyon MF (2000) LINE-1 elements and X chromosome inactivation: a function for “junk” DNA? Proc Natl Acad Sci USA 97:6248–6249
Madsen O, Scally M, Douady CJ, Kao DJ, DeBry RW, Adkins R, Amrine HM, Stanhope MJ, de Jong WW, Springer MS (2001) Parallel adaptive radiations in two major clades of placental mammals. Nature 409:610–614
Malik HS, Burke WD, Eickbush TH (1999) The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol 16:793–805
Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, Keith JC Jr, McCoy JM (2000) Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785–789
Murphy WJ, Eizirik E, Johnson WE, Zhang YP, Ryder OA, O’Brien SJ (2001a) Molecular phylogenetics and the origins of placental mammals. Nature 409:614–618
Murphy WJ, Eizirik E, O’Brien SJ, Madsen O, Scally M, Douady CJ, Teeling E, Ryder OA, Stanhope MJ, de Jong WW, Springer MS (2001b) Resolution of the early placental mammal radiation using bayesian phylogenetics. Science 294:2348–2351
Nekrutenko A, Li WH (2001) Transposable elements are found in a large number of human protein-coding genes. Trends Genet 17:619–621
Nikaido M, Nishihara H, Hukumoto Y, Okada N (2003) Ancient SINEs from African endemic mammals. Mol Biol Evol 20:522–527
Ovchinnikov I, Troxel AB, Swergold GD (2001) Genomic characterization of recent human LINE-1 insertions: evidence supporting random insertion. Genome Res 11:2050–2058
Parish DA, Vise P, Wichman HA, Bull JJ, Baker RJ (2002) Distribution of LINEs and other repetitive elements in the karyotype of the bat Carollia: implications for X-chromosome inactivation. Cytogenet Gen Res 96:191–197
Pascale E, Valle E, Furano AV (1990) Amplification of an ancestral mammalian L1 family of long interspersed repeated DNA occurred just before the murine radiation. Proc Natl Acad Sci USA 87:9481–9485
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Seabright M (1971) A rapid banding technique for human chromosomes. Lancet 2:971–972
Springer MS, Murphy WJ, Eizirik E, O’Brien SJ (2003) Placental diversification and the Cretaceous-Tertiary boundary. Proc Natl Acad Sci USA 100:1056–1061
Sumner A (1972) A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res 75:304–306
Thomsen PD, Miller JR (1996) Pig genome analysis: differential distribution of SINE and LINE sequences is less pronounced than in the human and mouse genomes. Mamm Genome 7:42–46
Usdin K, Chevret P, Catzeflis FM, Verona R, Furano AV (1995) L1 (LINE-1) retrotransposable elements provide a “fossil” record of the phylogenetic history of murid rodents. Mol Biol Evol 12:73–82
Verneau O, Catzeflis F, Furano AV (1998) Determining and dating recent rodent speciation events by using L1 (LINE-1) retrotransposons. Proc Natl Acad Sci USA 95:11284–11289
Volff JN, Korting C, Froschauer A, Sweeney K, Schartl M (2001) Non-LTR retrotransposons encoding a restriction enzyme-like endonuclease in vertebrates. J Mol Evol 52:351–360
Waddell PJ, Kishino H, Ota R (2001) A phylogenetic foundation for comparative mammalian genomics. Genome Inform Ser Workshop Genome Inform 12:141–154
Waddell PJ, Shelley S (2003) Evaluating placental inter-ordinal phylogenies with novel sequences including RAG1, gamma-fibrinogen, ND6, and mt-tRNA, plus MCMC-driven nucleotide, amino acid, and codon models. Mol Phylogenet Evol 28:197–224
Wichman HA, Van den Bussche RA, Hamilton MJ, Baker RJ (1992) Transposable elements and the evolution of genome organization in mammals. Genetica 86:287–293
Yang Z, Boffelli D, Boonmark N, Schwartz K, Lawn R (1998) Apolipoprotein(a) gene enhancer resides within a LINE element. J Biol Chem 273:891–897
Zhang J, Peterson T (1999) Genome rearrangements by nonlinear transposons in maize. Genetics 153:1403–1410
Acknowledgements
Funding from the South African National Research Foundation (GUN 2053812), the University of Stellenbosch and the Wellcome Trust is gratefully acknowledged. The authors are indebted to Vitaly Volobouev and Céline Canler, Muséum National d’Histoire Naturelle (Paris) for providing the anteater, sloth, hyaena, galago and springbok cell lines, and to Robert Bond (Florida Center for Aquatic Resources Studies) and Julio C. Pieczarka (Belem University, Brazil) for manatee and armadillo material, respectively.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Henikoff
Paul D. Waters and Gauthier Dobigny contributed equally to this work
Rights and permissions
About this article
Cite this article
Waters, P.D., Dobigny, G., Pardini, A.T. et al. LINE-1 distribution in Afrotheria and Xenarthra: implications for understanding the evolution of LINE-1 in eutherian genomes. Chromosoma 113, 137–144 (2004). https://doi.org/10.1007/s00412-004-0301-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-004-0301-9