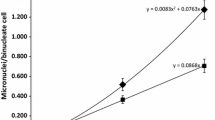
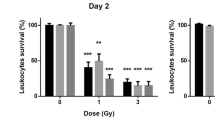
Abstract
At the Center for High-Throughput Minimally Invasive Radiation Biodosimetry, we have developed a rapid automated biodosimetry tool (RABiT); this is a completely automated, ultra-high-throughput robotically based biodosimetry workstation designed for use following a large-scale radiological event, to perform radiation biodosimetry measurements based on a fingerstick blood sample. High throughput is achieved through purpose built robotics, sample handling in filter-bottomed multi-well plates and innovations in high-speed imaging and analysis. Currently, we are adapting the RABiT technologies for use in laboratory settings, for applications in epidemiological and clinical studies. Our overall goal is to extend the RABiT system to directly measure the kinetics of DNA repair proteins. The design of the kinetic/time-dependent studies is based on repeated, automated sampling of lymphocytes from a central reservoir of cells housed in the RABiT incubator as a function of time after the irradiation challenge. In the present study, we have characterized the DNA repair kinetics of the following repair proteins: γ-H2AX, 53-BP1, ATM kinase, MDC1 at multiple times (0.5, 2, 4, 7 and 24 h) after irradiation with 4 Gy γ rays. In order to provide a consistent dose exposure at time zero, we have developed an automated capillary irradiator to introduce DNA DSBs into fingerstick-size blood samples within the RABiT. To demonstrate the scalability of the laboratory-based RABiT system, we have initiated a population study using γ-H2AX as a biomarker.




Similar content being viewed by others
References
Andegeko Y, Moyal L, Mittelman L, Tsarfaty I, Shiloh Y, Rotman G (2001) Nuclear retention of ATM at sites of DNA double strand breaks. J Biol Chem 276:38224–38230
Ang KK, Jiang GL, Guttenberger R, Thames HD, Stephens LC, Smith CD, Feng Y (1992) Impact of spinal cord repair kinetics on the practice of altered fractionation schedules. Radiother Oncol 25:287–294
Banath JP, Macphail SH, Olive PL (2004) Radiation sensitivity, H2AX phosphorylation, and kinetics of repair of DNA strand breaks in irradiated cervical cancer cell lines. Cancer Res 64:7144–7149
Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J (2005) Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J Cell Biol 170:201–211
Bhogal N, Kaspler P, Jalali F, Hyrien O, Chen R, Hill RP, Bristow RG (2010) Late residual gamma-H2AX foci in murine skin are dose responsive and predict radiosensitivity in vivo. Radiat Res 173:1–9
Bouquet F, Muller C, Salles B (2006) The loss of gammaH2AX signal is a marker of DNA double strand breaks repair only at low levels of DNA damage. Cell Cycle 5:1116–1122
Chen Y, Wang H, Garty G, Xu Y, Lyulko OV, Turner HC, Randers-Pehrson G, Simaan N, Yao YL, Brenner DJ (2009) Design and preliminary validation of a rapid automated biosodimetry tool for high throughput radiological triage. Proc ASME Des Eng Tech Conf 3:61–67
Chen Y, Wang H, Garty G, Xu Y, Lyulko OV, Turner HC, Randers-Pehrson G, Simaan N, Yao YL, Brenner DJ (2010) Development of a robotically based automated biodosimetry tool for high-throughput radiological triage. Int J Biomech Biomed Rob 1:115–125
Chen Y, Wang H, Zhang J, Garty G, Simaan N, Yao YL, Brenner DJ (2012) Automated recognition of robotic manipulation failures in high-throughput biodosimetry tool. Expert Syst Appl 39:9602–9611
Collins AR, Azqueta A (2012) DNA repair as a biomarker in human biomonitoring studies; further applications of the comet assay. Mutat Res 736:122–129
Costes SV, Chiolo I, Pluth JM, Barcellos-Hoff MH, Jakob B (2010) Spatiotemporal characterization of ionizing radiation induced DNA damage foci and their relation to chromatin organization. Mutat Res 704:78–87
Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N (2010) Computer control of microscopes using μManager. Curr Protocol Mol Biol 14.20.1–14.20.17
Fenech M (2010) The lymphocyte cytokinesis-block micronucleus cytome assay and its application in radiation biodosimetry. Health Phys 98:234–243
Fenech M, Chang WP, Kirsch-Volders M, Holland N, Bonassi S, Zeiger E (2003) HUMN project: detailed description of the scoring criteria for the cytokinesis-block micronucleus assay using isolated human lymphocyte cultures. Mutat Res 534:65–75
Frankenberg-Schwager M (1989) Review of repair kinetics for DNA damage induced in eukaryotic cells in vitro by ionizing radiation. Radiother Oncol 14:307–320
Garty G, Chen Y, Salerno A, Turner HC, Zhang J, Lyulko O, Bertucci A, Xu Y, Wang H, Simaan N, Randers-Pehrson G, Yao YL, Amundson SA, Brenner DJ (2010) The RABiT: a rapid automated biodosimetry tool for radiological triage. Health Phys 68:209–217
Garty G, Chen Y, Turner HC, Zhang J, Lyulko O, Bertucci A, Xu Y, Wang H, Simaan N, Randers-Pehrson G, Yao YL, Brenner DJ (2011) The RABIT: a rapid automated biodosimetry tool for radiological triage. II technological developments. Int J Radiat Biol 87:776–790
Hable V, Drexler GA, Brüning T, Burgdorf C, Greubel C, Derer A, Seel J, Strickfaden H, Cremer T, Friedl AA, Dollinger G (2012) Recruitment kinetics of DNA repair proteins Mdc1 and Rad52 but not 53BP1 depend on damage complexity. PLoS ONE 7:1–11
Ivashkevich A, Redon CE, Nakamura AJ, Martin RF, Martin OA (2012) Use of the γ-H2AX assay to monitor DNA damage and repair in translational cancer research. Cancer Lett 327:123–133
Jungmichel S, Stucki M (2010) MDC1: the art of keeping things in focus. Chromosoma 119:337–349
Kinner A, Wu W, Staudt C, Iliakis G (2008) Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res 36:5678–5694
Kuhne M, Riballo E, Rief N, Rothkamm K, Jeggo PA, Lobrich M (2004) A double-strand break repair defect in ATM-deficient cells contributes to radiosensitivity. Cancer Res 64:500–508
Löbrich M, Rief N, Kuhne M, Heckmann M, Fleckenstein J, Rube C, Uder M (2005) In vivo formation and repair of DNA double-strand breaks after computed tomography examinations. Proc Natl Acad Sci USA 102:8984–8989
Lyulko OV, Garty G, Randers-Pehrson G, Turner HC and Brenner DJ (2014) Fast image analysis for the micronucleus assay in a fully automated high throughput biodosimetry system. Radiat Res (accepted)
MacPhail SH, Banath JP, Yu TY et al (2003) Expression of phosphorylated histone H2AX in cultured cell lines following exposure to X-rays. Int J Radiat Biol 9:351–358
Markova E, Schultz N, Belyaev IY (2007) Kinetics and dose-response of residual 53BP1/gamma-H2AX foci: co-localization, relationship with DSB repair and clonogenic survival. Int J Radiat Biol 83:319–329
Martin NT, Nahas SA, Tunuguntla R, Fike F, Gatti RA (2011) Assessing ‘radiosensitivity’ with kinetic profiles of γ-H2AX, 53BP1 and BRCA1 foci. Radiother Oncol 101:35–38
Metzger L, Iliakis G (1991) Kinetics of DNA double-strand break repair throughout the cell cycle as assayed by pulsed field gel electrophoresis in CHO cells. Int J Radiat Biol 59:1325–1339
Nakamura A, Sedelnikova OA, Redon C, Pilch DR, Sinogeeva NI, Shroff R, Lichten M, Bonner WM (2006) Techniques for gamma-H2AX detection. Methods Enzymol 409:236–250
Rasband WS (1997-2012) ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA. http://imagej.nih.gov/ij/
Redon CE, Dickey JS, Bonner WM, Sedelnikova OA (2009) gamma-H2AX as a biomarker of DNA damage induced by ionizing radiation in human peripheral blood lymphocytes and artificial skin. Advan Space Res 43:1171–1178
Rogakou EP, Pilch PR, Orr AH, Ivanova VS, Bonner WM (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273:5858–5868
Rothkamm K, Horn S (2009) gamma-H2AX as protein biomarker for radiation exposure. Ann Ist Super Sanita 45:265–271
Rothkamm K, Löbrich M (2003) Evidence for a lack of DNA double-strand break repair in human cells exposed to very low X-ray doses. Proc Natl Acad Sci USA 100:5057–5062
Rube CE, Grudzenski S, Kuhne M, Dong X, Rief N, Lobrich M, Rube C (2008) DNA double-strand break repair of blood lymphocytes and normal tissues analysed in a preclinical mouse model: implications for radiosensitivity testing. Clin Cancer Res 14:6546–6555
Schultz LB, Chehab NH, Malikzay A, Halazonetis TD (2000) p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol 151:1381–1390
Sedelnikova OA, Pilch DR, Redon C, Bonner WM (2003) Histone H2AX in DNA damage and repair. Cancer Biol Ther 2:233–235
Taneja N, Davis M, Choy JS et al (2004) Histone H2AX phosphorylation as a predictor of radiosensitivity and target for radiotherapy. J Biol Chem 279:2273–2280
Turner HC, Brenner DJ, Chen Y, Bertucci A, Zhang J, Wang H, Lyulko OV, Xu Y, Shuryak I, Schaefer J, Simaan N, Randers-Pehrson G, Yao YL, Amundson SA, Garty G (2011) Adapting the γ-H2AX assay for automated processing in human lymphocytes. 1 technological aspects. Radiat Res 175:282–290
Ugenskiene R, Prise K, Folkard M, Lekki J, Stachura Z, Zazula M, Stachura J (2009) Dose response and kinetics of foci disappearance following exposure to high- and low-LET ionizing radiation. Int J Radiat Biol 85:872–882
Van den Aardweg GJ, Hopewell JW, Guttenberger R (1996) The kinetics of repair of sublethal radiation-induced damage in pig skin: studies with multiple interfraction intervals. Radiat Res 145:586–594
Wilson PF, Nham PB, Urbin SS, Hinz JM, Jones IM, Thompson LH (2010) Inter-individual variation in DNA double-strand break repair in human fibroblasts before and after exposure to low doses of ionizing radiation. Mutat Res 683:91–97
Xu Y, Turner HC, Garty G, Brenner DJ (2013) A rapid, quantitative method to characterize the human lymphocyte concentration for automated high-throughput radiation biodosimetry. Biomed Eng Res 2:16–19
Acknowledgments
This work was supported by Grant Numbers U19-AI067773 and 1R21-ES019494, for the Center for High-Throughput Minimally Invasive Radiation Biodosimetry, from the National Institute of Allergy and Infectious Diseases, National Institute of Environmental Health Sciences and National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases, National Institute of Environmental Health Sciences or the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Turner, H.C., Sharma, P., Perrier, J.R. et al. The RABiT: high-throughput technology for assessing global DSB repair. Radiat Environ Biophys 53, 265–272 (2014). https://doi.org/10.1007/s00411-014-0514-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00411-014-0514-0