Abstract
Background
Head and neck squamous cell carcinoma (HNSCC) is a heterogeneous disease characterized by different molecular subtypes with different prognosis and response to treatment. Therefore, the aim of this study was to construct reliable gene signatures based on immune checkpoint-related genes to distinguish between subgroups of patients with different risks.
Methods
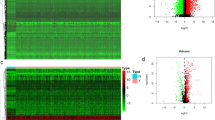
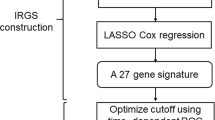
We obtained the HNSCC data from The Cancer Genome Atlas (TCGA) database and Gene Expression Omnibus (GEO) as a training set and the external validation set, respectively. First, differentially expressed immune checkpoint-related genes in tumor tissues and normal tissues were determined, and the potential functions of differential genes were explored through GO function annotation and KEGG pathway enrichment analysis. Using univariate Cox regression analysis, 20 immune checkpoint-related genes in HNSCC patients were significantly associated with overall survival (OS). Subsequently, seven genes were selected by multivariate Cox regression analysis to create a gene signature. Next, the stability of gene signatures was assessed using Kaplan–Meier curve, Time-dependent receiver operating characteristic (ROC) curve. Finally, we constructed a nomogram visualization modelled to facilitate subsequent clinical applications.
Results
A total of 80 differentially expressed genes (DEGs) were obtained, the GO analysis of these DEGs indicated that they were significantly enriched in positive regulation of cell activation, T cell activation; the KEGG analysis results performed and showed that the DEGs were enriched in the MAPK signaling pathway, PI3K − Akt signaling pathway. 7 genes (PPP2R1B, MYD88, CD86, CD80, MAP2K1, TRIB3 and ICOS) were screened by univariate and multivariate Cox regression, and they were used to construct a prognostic model. In the TCGA and GEO datasets, Kaplan–Meier analysis indicated that patients in the high-risk group have a poor prognosis. The sensitivity and specificity evaluation of prognostic model for 1-, 3-, 5-year OS in TCGA were 0.644, 0.661 and 0.625, respectively; and in GSE41613 were 0.748, 0.719, and 0.727, respectively. The calibration chart curve showed that the nomogram has strong clinical performance in the prognosis prediction of HNSCC patients.
Conclusions
A novel immune checkpoint-related gene signature can effectively predict and stratify OS in HNSCC patients.








Similar content being viewed by others
Availability of data and materials
The raw data of this study are derived from the TCGA database (https://portal.gdc.cancer.gov/) and GEO data portal (https://www.ncbi.nlm.nih.gov/geo/), which are publicly available databases.
References
Kobayashi K, Hisamatsu K, Suzui N, Hara A, Tomita H, Miyazaki T (2018) A review of HPV-related head and neck cancer. J Clin Med 7:9. https://doi.org/10.3390/jcm7090241
Chow LQM (2020) Head and neck cancer. New England J Med 382(1):60–72. https://doi.org/10.1056/NEJMra1715715
Argiris A, Karamouzis MV, Raben D, Ferris RL (2008) Head and neck cancer. Lancet (London, England) 371(9625):1695–1709. https://doi.org/10.1016/s0140-6736(08)60728-x
Siegel RL, Miller KD, Jemal A (2017) Cancer statistics. CA Cancer J Clin 67(1):7–30. https://doi.org/10.3322/caac.21387
Meucci S, Keilholz U, Tinhofer I, Ebner OA (2016) Mutational load and mutational patterns in relation to age in head and neck cancer. Oncotarget 7(43):69188–69199. https://doi.org/10.18632/oncotarget.11312
Mandal R, Şenbabaoğlu Y, Desrichard A, Havel JJ, Dalin MG, Riaz N, Lee KW, Ganly I, Hakimi AA, Chan TA, Morris LG (2016) The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight 1(17):e89829. https://doi.org/10.1172/jci.insight.89829
Mehra R, Seiwert TY, Gupta S, Weiss J, Gluck I, Eder JP, Burtness B, Tahara M, Keam B, Kang H, Muro K, Geva R, Chung HC, Lin CC, Aurora-Garg D, Ray A, Pathiraja K, Cheng J, Chow LQM, Haddad R (2018) Efficacy and safety of pembrolizumab in recurrent/metastatic head and neck squamous cell carcinoma: pooled analyses after long-term follow-up in KEYNOTE-012. Br J Cancer 119(2):153–159. https://doi.org/10.1038/s41416-018-0131-9
Lydiatt WM, Patel SG, Sullivan B, Brandwein MS, Ridge JA, Migliacci JC, Loomis AM, Shah JP (2017) Head and Neck cancers-major changes in the American Joint Committee on cancer eighth edition cancer staging manual. CA Cancer J Clin 67(2):122–137
Bobdey S, Mair M, Nair S, Nair D, Balasubramaniam G, Chaturvedi P (2018) A Nomogram based prognostic score that is superior to conventional TNM staging in predicting outcome of surgically treated T4 buccal mucosa cancer: time to think beyond TNM. Oral Oncol 81:10–15. https://doi.org/10.1016/j.oraloncology.2018.04.002
Almendro V, Marusyk A, Polyak K (2013) Cellular heterogeneity and molecular evolution in cancer. Ann Rev Pathol 8:277–302. https://doi.org/10.1146/annurev-pathol-020712-163923
Mroz EA, Tward AD, Hammon RJ, Ren Y, Rocco JW (2015) Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med 12(2):e1001786. https://doi.org/10.1371/journal.pmed.1001786
Liu F, Huang J, He F, Ma X, Fan F, Meng M, Zhuo Y, Zhang L (2020) CD96, a new immune checkpoint, correlates with immune profile and clinical outcome of glioma. Scient Rep 10(1):10768. https://doi.org/10.1038/s41598-020-66806-z
Zhang Z, Liu J, Zhang C, Li F, Li L, Wang D, Chand D, Guan F, Zang X, Zhang Y (2020) Over-expression and prognostic significance of HHLA2, a new immune checkpoint molecule, in human clear cell renal cell carcinoma. Front Cell Dev Biol 8:280. https://doi.org/10.3389/fcell.2020.00280
Scandiuzzi L, Ghosh K, Zang X (2011) T cell costimulation and coinhibition: genetics and disease. Discov Med 12(63):119–128
Shan N, Li N, Dai Q, Hou L, Yan X, Amei A, Lu L, Wang Z (2020) Interplay of tRNA-derived fragments and T cell activation in breast cancer patient survival. Cancers 12:8. https://doi.org/10.3390/cancers12082230
Xie X, Xiong G, Wang Q, Ge Y, Cui X (2019) Long non-coding RNA LINC00460 promotes head and neck squamous cell carcinoma cell progression by sponging miR-612 to up-regulate AKT2. Am J Trans Res 11(10):6326–6340
Zhao ZJ, Liu FY, Li P, Ding X, Zong ZH, Sun CF (2011) CCL19-induced chemokine receptor 7 activates the phosphoinositide-3 kinase-mediated invasive pathway through Cdc42 in metastatic squamous cell carcinoma of the head and neck. Oncol Rep 25(3):729–737. https://doi.org/10.3892/or.2010.1109
Ji X, Guo X, Wang Y, Li X, Li H (2020) Rab18 regulates proliferation, invasion and cisplatin sensitivity through STAT3 signaling in head and neck squamous cell carcinoma. OncoTargets Thera 13:4123–4134. https://doi.org/10.2147/ott.S238503
Chen L, Li YC, Wu L, Yu GT, Zhang WF, Huang CF, Sun ZJ (2018) TRAF6 regulates tumour metastasis through EMT and CSC phenotypes in head and neck squamous cell carcinoma. J Cell Mol Med 22(2):1337–1349. https://doi.org/10.1111/jcmm.13439
Ju H, Hu Z, Lu Y, Wu Y, Zhang L, Wei D, Guo W, Xia W, Liu S, Ren G, Hu J (2020) TLR4 activation leads to anti-EGFR therapy resistance in head and neck squamous cell carcinoma. Am J Cancer Res 10(2):454–472
García-Escudero R, Segrelles C, Dueñas M, Pombo M, Ballestín C, Alonso-Riaño M, Nenclares P, Álvarez-Rodríguez R, Sánchez-Aniceto G, Ruíz-Alonso A, López-Cedrún JL, Paramio JM, Lorz C (2018) Overexpression of PIK3CA in head and neck squamous cell carcinoma is associated with poor outcome and activation of the YAP pathway. Oral Oncol 79:55–63. https://doi.org/10.1016/j.oraloncology.2018.02.014
El Taweel M, Gawdat RM, Abdelfattah R (2020) Prognostic Impact of PPP2R5C Gene Expression in Adult Acute Myeloid Leukemia Patients with Normal Cytogenetics. Indian J Hematol Blood Trans Soc 36(1):37–46. https://doi.org/10.1007/s12288-019-01142-5
Tian H, Yin L, Ding K, Xia YY, Wang XH, Wu JZ, He X (2018) Raf1 is a prognostic factor for progression in patients with non-small cell lung cancer after radiotherapy. Oncol Rep 39(4):1966–1974. https://doi.org/10.3892/or.2018.6277
Hu JW, Ding GY, Fu PY, Tang WG, Sun QM, Zhu XD, Shen YH, Zhou J, Fan J, Sun HC, Huang C (2020) Identification of FOS as a candidate risk gene for liver cancer by integrated bioinformatic analysis. Biomed Res Int 20:6784138. https://doi.org/10.1155/2020/6784138
Zhou Y, Chen P, Huang Q, Wan T, Jiang Y, Jiang S, Yan S, Zheng M (2020) Overexpression of YES1 is associated with favorable prognosis and increased platinum-sensitivity in patients with epithelial ovarian cancer. Histol Histopathol 35(7):721–728. https://doi.org/10.14670/hh-18-203
Wang SS, Esplin ED, Li JL, Huang L, Gazdar A, Minna J, Evans GA (1998) Alterations of the PPP2R1B gene in human lung and colon cancer. Science (New York, NY) 282(5387):284–287. https://doi.org/10.1126/science.282.5387.284
Koch AT, Love-Homan L, Espinosa-Cotton M, Stanam A, Simons AL (2015) MyD88-dependent signaling decreases the antitumor efficacy of epidermal growth factor receptor inhibition in head and neck cancer cells. Can Res 75(8):1657–1667. https://doi.org/10.1158/0008-5472.Can-14-2061
Mansour A, Elkhodary T, Darwish A, Mabed M (2014) Increased expression of costimulatory molecules CD86 and sCTLA-4 in patients with acute lymphoblastic leukemia. Leukemia Lymphoma 55(9):2120–2124. https://doi.org/10.3109/10428194.2013.869328
Feng XY, Lu L, Wang KF, Zhu BY, Wen XZ, Peng RQ, Ding Y, Li DD, Li JJ, Li Y, Zhang XS (2019) Low expression of CD80 predicts for poor prognosis in patients with gastric adenocarcinoma. Future Oncol (London, England) 15(5):473–483. https://doi.org/10.2217/fon-2018-0420
Huynh H, Nguyen TT, Chow KH, Tan PH, Soo KC, Tran E (2003) Over-expression of the mitogen-activated protein kinase (MAPK) kinase (MEK)-MAPK in hepatocellular carcinoma: its role in tumor progression and apoptosis. BMC Gastroenterol 3:19. https://doi.org/10.1186/1471-230x-3-19
Wang XJ, Li FF, Zhang YJ, Jiang M, Ren WH (2020) TRIB3 promotes hepatocellular carcinoma growth and predicts poor prognosis. Cancer Bio Section A Dis Markers. https://doi.org/10.3233/cbm-201577
Zhang Y, Luo Y, Qin SL, Mu YF, Qi Y, Yu MH, Zhong M (2016) The clinical impact of ICOS signal in colorectal cancer patients. Oncoimmunology 5(5):e1141857. https://doi.org/10.1080/2162402x.2016.1141857
Funding
None.
Author information
Authors and Affiliations
Contributions
This research was conducted in collaboration with all authors. DS and JT performed the data curation and analysis. DS, JT and XL analyzed and interpreted the results. DS, JT and XH drafted and reviewed the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval
No permissions were required to use the repository data.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Song, D., Tian, J., Han, X. et al. A model of seven immune checkpoint-related genes predicting overall survival for head and neck squamous cell carcinoma. Eur Arch Otorhinolaryngol 278, 3467–3477 (2021). https://doi.org/10.1007/s00405-020-06540-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00405-020-06540-4