Abstract
Purpose
The aim of the study is to identify a reliable gene panel to predict the prognosis of HNSCC patients by integrated genomic analysis.
Methods
Co-expression gene networks were constructed by WGCNA using GSE113282 gene expression profile. The biological functional investigation was performed by GO and KEGG function enrichment analysis. The hub gene module was screened by PPI. The prognostic gene panel was established by Lasso regression analysis, and further progression-free survival (PFS) analysis was validated by Kaplan–Meier survival analysis using GSE102995 data.
Results
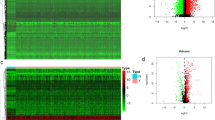
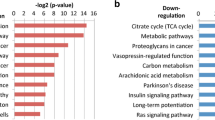
We identified 195 genes associated with the overall survival (OS) status (correlation coefficients: − 0.42, and p value: 2e−05) by WGCNA. These genes were enriched in immune-related cytokines and pathways analyzed by GO and KEGG. Among the 195 genes, the module (42 genes) with the highest score was screened by PPI. A novel seven-gene predictive panel (CD19, CD40LG, CD5, CXCR6, FPR2, NCAM1, and SELL) was established by Lasso regression analysis, and the area under ROC curve (AUC) for 3-year OS status was 0.8298 and 0.7571, respectively, in the training set and the test set. The PFS time of the low-risk patients was significantly longer than the high-risk patients (p < 0.0001; log-rank test) by further validation using GSE102995 data.
Conclusion
The seven-gene panel may serve as a reliable predictive tool for HNSCC patients treated with platinum-based radio (chemo) therapy, and may be potential therapeutic targets for HNSCC patients.





Similar content being viewed by others
Code availability
The data in our study are available from the corresponding author upon reasonable request.
Abbreviations
- SCC:
-
Squamous cell carcinoma
- HNSCC:
-
Head and neck squamous cell carcinoma
- NCBI:
-
National Center Biotechnology Information
- GEO:
-
Gene Expression Omnibus
- OPSCC:
-
Oropharyngeal squamous cell carcinoma
- WGCNA:
-
Weighted correlation network analysis
- OS:
-
Overall survival
- GO:
-
Gene ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- BP:
-
Biological process
- CC:
-
Cellular component
- MF:
-
Molecular function
- STRING:
-
The Search Tool for the Retrieval of Interacting Genes/Proteins
- PPI:
-
Protein–protein interaction
- MCODE:
-
Molecular Complex Detection
- ROC:
-
Receiver operating characteristic
- Lasso:
-
Least absolute shrinkage and selection operator
- AUC:
-
Area under curve
- PFS:
-
Progression-free survival
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424. https://doi.org/10.3322/caac.21492
Argiris A, Karamouzis MV, Raben D, Ferris RL (2008) Head and neck cancer. Lancet 371(9625):1695–1709. https://doi.org/10.1016/S0140-6736(08)60728-X
Kaidar-Person O, Gil Z, Billan S (2018) Precision medicine in head and neck cancer. Drug Resist Updat 40:13–16. https://doi.org/10.1016/j.drup.2018.09.001
Ferris RL (2015) Immunology and Immunotherapy of Head and Neck Cancer. J Clin Oncol 33(29):3293–3304. https://doi.org/10.1200/JCO.2015.61.1509
Hess AK, Jöhrens K, Zakarneh A, Balermpas P, Von Der Grün J, Rödel C, Weichert W, Hummel M, Keilholz U, Budach V, Tinhofer I (2019) Characterization of the tumor immune micromilieu and its interference with outcome after concurrent chemoradiation in patients with oropharyngeal carcinomas. Oncoimmunology 8(8):1614858. https://doi.org/10.1080/2162402X.2019.1614858
Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Muertter RN, Edgar R (2009) NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 37:D885-890. https://doi.org/10.1093/nar/gkn764
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47. https://doi.org/10.1093/nar/gkv007
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinform 9:559. https://doi.org/10.1186/1471-2105-9-559
The Gene Ontology Consortium (2017) Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res 45(D1):D331–D338. https://doi.org/10.1093/nar/gkw1108
Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45(D1):D353–D361. https://doi.org/10.1093/nar/gkw1092
Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16(5):284–287. https://doi.org/10.1089/omi.2011.0118
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47(D1):D607–D613. https://doi.org/10.1093/nar/gky1131
Kohl M, Wiese S, Warscheid B (2011) Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol 696:291–303. https://doi.org/10.1007/978-1-60761-987-1_18
Friedman J, Hastie T, Tibshirani R (2010) Regularization paths for generalized linear models via coordinate descent. J Stat Softw 33(1):1–22
Siano M, Espeli V, Mach N, Bossi P, Licitra L, Ghielmini M, Frattini M, Canevari S, De Cecco L (2018) Gene signatures and expression of miRNAs associated with efficacy of panitumumab in a head and neck cancer phase II trial. Oral Oncol 82:144–151. https://doi.org/10.1016/j.oraloncology.2018.05.013
Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A (2016) Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin 66(4):271–289. https://doi.org/10.3322/caac.21349
Ong CJ, Shannon NB, Mueller S, Lek SM, Qiu X, Chong FT, Li K, Koh KKN, Tay GCA, Skanthakumar T, Hwang JSG, HonLim TK, Ang MK, Tan DSW, Tan NC, Tan HK, Soo KC, Iyer NG (2017) A three gene immunohistochemical panel serves as an adjunct to clinical staging of patients with head and neck cancer. Oncotarget. 8(45):79556–79566. https://doi.org/10.18632/oncotarget.18568
Liang LB, Huang XY, He H, Liu JY (2020) Prognostic values of radiosensitivity genes and CD19 status in gastric cancer: a retrospective study using TCGA database. Pharmgenomics Pers Med 13:365–373. https://doi.org/10.2147/PGPM.S265121
Korniluk A, Kemona H, Dymicka-Piekarska V (2014) Multifunctional CD40L: pro- and anti-neoplastic activity. Tumour Biol 35(10):9447–9457. https://doi.org/10.1007/s13277-014-2407-x
Rah YC, Ahn JC, Jeon EH, Kim H, Paik JH, Jeong WJ, Kwon SY, Ahn SH (2018) Low expression of CD40L in tumor-free lymph node of oral cavity cancer related with poor prognosis. Int J Clin Oncol 23(5):851–859. https://doi.org/10.1007/s10147-018-1294-3
Moreno-Manuel A, Jantus-Lewintre E, Simões I, Aranda F, Calabuig-Fariñas S, Carreras E, Zúñiga S, Saenger Y, Rosell R, Camps C, Lozano F, Sirera R (2020) CD5 and CD6 as immunoregulatory biomarkers in non-small cell lung cancer. Transl Lung Cancer Res. 9(4):1074–1083. https://doi.org/10.21037/tlcr-19-445
Mir H, Kaur G, Kapur N, Bae S, Lillard JW Jr, Singh S (2019) Higher CXCL16 exodomain is associated with aggressive ovarian cancer and promotes the disease by CXCR6 activation and MMP modulation. Sci Rep 9(1):2527. https://doi.org/10.1038/s41598-019-38766-6
Ikeda T, Nishita M, Hoshi K, Honda T, Kakeji Y, Minami Y (2020) Mesenchymal stem cell-derived CXCL16 promotes progression of gastric cancer cells by STAT3-mediated expression of Ror1. Cancer Sci 111(4):1254–1265. https://doi.org/10.1111/cas.14339
Kapur N, Mir H, Sonpavde GP, Jain S, Bae S, Lillard JW Jr, Singh S (2019) Prostate cancer cells hyper-activate CXCR6 signaling by cleaving CXCL16 to overcome effect of docetaxel. Cancer Lett 454:1–13. https://doi.org/10.1016/j.canlet.2019.04.001
Ke C, Ren Y, Lv L, Hu W, Zhou W (2017) Association between CXCL16/CXCR6 expression and the clinicopathological features of patients with non-small cell lung cancer. Oncol Lett 13(6):4661–4668. https://doi.org/10.3892/ol.2017.6088
Gastardelo TS, Cunha BR, Raposo LS, Maniglia JV, Cury PM, Lisoni FC, Tajara EH, Oliani SM (2014) Inflammation and cancer: role of annexin A1 and FPR2/ALX in proliferation and metastasis in human laryngeal squamous cell carcinoma. PLoS ONE 9(12):e111317. https://doi.org/10.1371/journal.pone.0111317
Lu J, Zhao J, Jia C, Zhou L, Cai Y, Ni J, Ma J, Zheng M, Lu A (2019) FPR2 enhances colorectal cancer progression by promoting EMT process. Neoplasma 66(5):785–791. https://doi.org/10.4149/neo_2018_181123N890
Shi Y, Wang J, Xin Z, Duan Z, Wang G, Li F (2015) Transcription factors and microRNA-co-regulated genes in gastric cancer invasion in ex vivo. PLoS ONE 10(4):e0122882. https://doi.org/10.1371/journal.pone.0122882
Ghaderi F, Ahmadvand S, Ramezani A, Montazer M, Ghaderi A (2018) Production and characterization of monoclonal antibody against a triple negative breast cancer cell line. Biochem Biophys Res Commun 505(1):181–186. https://doi.org/10.1016/j.bbrc.2018.09.087
Phadke GS, Satterwhite-Warden JE, Choudhary D, Taylor JA, Rusling JF (2018) A novel and accurate microfluidic assay of CD62L in bladder cancer serum samples. Analyst 143(22):5505–5511. https://doi.org/10.1039/c8an01463a
Choudhary D, Hegde P, Voznesensky O, Choudhary S, Kopsiaftis S, Claffey KP, Pilbeam CC, Taylor JA 3rd (2015) Increased expression of L-selectin (CD62L) in high-grade urothelial carcinoma: a potential marker for metastatic disease. Urol Oncol 33(9):387.e17–27. https://doi.org/10.1016/j.urolonc.2014.12.009
Funding
This work was supported by the Beijing Natural Science Foundation Program and Scientific Research Key Program of Beijing Municipal Commission of Education (KZ201910025034), the Beijing Municipal Administration of Hospitals’ Ascent Plan (DFL20180202), the National Key R&D Program of China (No. 2020YFB1312805), the Capital Health Research and Development of Special (No.2018–2-2054) and Beijing Municipal Administration of Hospitals Incubating Program” (PX2021008).
Author information
Authors and Affiliations
Contributions
LWW performed comparative analysis using bioinformatics tools. LWW, YFY, LF, CT, HZM, SZH, ML, RW and JGF participated in data analysis and discussion, LWW, RW and JGF interpreted data and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethics approval
For human datasets mentioned in this study, please refer to the original article (PMCID: PMC6682352 and PMID: 29909889). We re-analyzed the open accessed datasets, and the study was approved by the local ethics committees.
Availability of data and material
All data generated or analyzed during this study are included in this published article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, L., Yang, Y., Feng, L. et al. A novel seven-gene panel predicts the sensitivity and prognosis of head and neck squamous cell carcinoma treated with platinum-based radio(chemo)therapy. Eur Arch Otorhinolaryngol 278, 3523–3531 (2021). https://doi.org/10.1007/s00405-021-06717-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00405-021-06717-5