Abstract
Developmental and/or epileptic encephalopathies (DEEs) are a group of devastating genetic disorders, resulting in early-onset, therapy-resistant seizures and developmental delay. Here we report on 22 individuals from 15 families presenting with a severe form of intractable epilepsy, severe developmental delay, progressive microcephaly, visual disturbance and similar minor dysmorphisms. Whole exome sequencing identified a recurrent, homozygous variant (chr2:64083454A > G) in the essential UDP-glucose pyrophosphorylase (UGP2) gene in all probands. This rare variant results in a tolerable Met12Val missense change of the longer UGP2 protein isoform but causes a disruption of the start codon of the shorter isoform, which is predominant in brain. We show that the absence of the shorter isoform leads to a reduction of functional UGP2 enzyme in neural stem cells, leading to altered glycogen metabolism, upregulated unfolded protein response and premature neuronal differentiation, as modeled during pluripotent stem cell differentiation in vitro. In contrast, the complete lack of all UGP2 isoforms leads to differentiation defects in multiple lineages in human cells. Reduced expression of Ugp2a/Ugp2b in vivo in zebrafish mimics visual disturbance and mutant animals show a behavioral phenotype. Our study identifies a recurrent start codon mutation in UGP2 as a cause of a novel autosomal recessive DEE syndrome. Importantly, it also shows that isoform-specific start-loss mutations causing expression loss of a tissue-relevant isoform of an essential protein can cause a genetic disease, even when an organism-wide protein absence is incompatible with life. We provide additional examples where a similar disease mechanism applies.
Similar content being viewed by others
Introduction
Developmental and/or epileptic encephalopathies (DEEs) are a heterogeneous group of genetic disorders, characterized by severe epileptic seizures in combination with developmental delay or regression [49]. Genes involved in multiple pathophysiological pathways have been implicated in DEEs, including synaptic impairment, ion channel alterations, transporter defects and metabolic processes such as disorders of glycosylation [68]. Mostly, dominant acting, de novo mutations have been identified in children suffering from DEEs [32], and only a limited number of genes with a recessive mode of inheritance are known so far, with a higher occurrence rate in consanguineous populations [71]. A recent cohort study on DEEs employing whole exome sequencing (WES) and copy number analysis, however, found that up to 38% of diagnosed cases might be caused by recessive genes, indicating that the importance of this mode of inheritance in DEEs has been underestimated [74].
The human genome contains ~ 20,000 genes of which more than 5000 have been implicated in genetic disorders. Wide-scale population genomic studies and CRISPR–Cas9-based loss-of-function (LoF) screens have identified around 3000–7000 genes that are essential for the viability of the human organism or result in profound loss of fitness when mutated, in agreement with that they are depleted for LoF variants in the human population [10]. For some of these essential genes, it is believed that LoF variants are incompatible with life and are, therefore, unlikely to be implicated in genetic disorders presenting in postnatal life [84]. One such example is the UDP-glucose pyrophosphorylase (UGP2) gene at chromosome 2. UGP2 is an essential octameric enzyme in nucleotide sugar metabolism [38, 39, 121], as it is the only known enzyme capable of catalyzing the conversion of glucose-1-phosphate to UDP-glucose [36, 108]. UDP-glucose is a crucial precursor for the production of glycogen by glycogen synthase (GYS) [2, 44], and also serves as a substrate for UDP-glucose:glycoprotein transferases (UGGT) and UDP-glucose-6-dehydrogenase (UGDH), thereby playing important roles in glycoprotein folding control, glycoconjugation and UDP-glucuronic acid synthesis. The latter is an obligate precursor for the synthesis of glycosaminoglycans and proteoglycans of the extracellular matrix [65, 110], of which aberrations have been associated with DEEs and neurological disorders [4, 24, 77, 98]. UGP2 has previously been identified as a marker protein in various types of malignancies including gliomas where its upregulation is correlated with a poor disease outcome [27, 59, 61, 101, 103, 111, 112, 122], but has so far not been implicated in genetic diseases and it has been speculated that this is given its essential role in metabolism [38].
Many genes are differentially expressed amongst tissues, regulated by non-coding regulatory elements [76]. In addition, it has become clear that there are more than 40,000 protein isoforms encoded in the human genome, whose expression levels vary amongst tissues. Although there are examples of genetic disorders caused by the loss of tissue-specific protein isoforms [41, 47, 57, 100], it is unknown whether a tissue-relevant loss of an essential gene can be involved in human disease. Here, we report on such a scenario, providing evidence that a novel form of a severe DEE syndrome is caused by the brain-relevant loss of the essential gene UGP2 due to an isoform-specific and germ line-transmitted start codon mutation. We present data that this is likely a more frequent disease mechanism in human genetics, illustrating that essential genes for which organism-wide loss is lethal can still be implicated in genetic disease when only absent in certain tissues due to expression misregulation.
Methods
Patient recruitment
All affected probands were investigated by their referring physicians and all genetic analyses were performed in a diagnostic setting. Legal guardians of affected probands gave informed consent for genomic investigations and publication of their anonymized data.
Next-generation sequencing of index patients
Individual 1
Genomic DNA was isolated from peripheral blood leukocytes of the proband and both parents, and exome-coding DNA was captured with the Agilent SureSelect Clinical Research Exome (CRE) kit (v2). Sequencing was performed on an Illumina HiSeq 4000 with 150-bp paired-end reads. Reads were aligned to hg19 using BWA (BWA-MEM v0.7.13) and variants were called using the GATK haplotype caller (v3.7 (reference: https://www.broadinstitute.org/gatk/) [67]. Detected variants were annotated, filtered and prioritized using the Bench lab NGS v5.0.2 platform (Agilent technologies). Initially, only genes known to be involved in epilepsy were analyzed, followed by a full exome analysis revealing the homozygous UGP2 variant.
Individuals 2, 3 and 4
Using genomic DNA from the proband and parents (individual 4) or the proband, parents, and affected sibling (individuals 2 and 3), the exonic regions and flanking splice junctions of the genome were captured using the SureSelect Human All Exon V4 (50 Mb) (individual 4) or the IDT xGen Exome Research Panel v1.0 (individuals 2 and 3). Massively parallel (NextGen) sequencing was done on an Illumina system with 100 bp or greater paired-end reads. Reads were aligned to human genome build GRCh37/UCSC hg19 and analyzed for sequence variants using a custom-developed analysis tool. Additional sequencing technology and variant interpretation protocol has been previously described [82]. The general assertion criteria for variant classification are publicly available on the GeneDx ClinVar submission page (https://www.ncbi.nlm.nih.gov/clinvar/submitters/26957/).
Individual 5
Diagnostic exome sequencing was done at the Departments of Human Genetics of the Radboud University Medical Center Nijmegen, The Netherlands, and performed essentially as described previously [96].
Individuals 6, 7, 8, 9, 10, 14, 15, 16, 17 and 18
After informed consent, we collected blood samples from the probands, their parents and unaffected siblings, and extracted DNA using standard procedures. To investigate the genetic cause of the disease, WES was performed in the affected proband. Nextera Rapid Capture Enrichment kit (Illumina) was used according to the manufacturer’s instructions. Libraries were sequenced in an Illumina HiSeq3000 using a 100-bp paired-end reads protocol. Sequence alignment to the human reference genome (UCSC hg19), variants calling, and annotation were performed as described elsewhere [69]. After removing all synonymous changes, we filtered single nucleotide variants (SNVs) and indels, only considering exonic and donor/acceptor splicing variants. In accordance with the pedigree and phenotype, priority was given to rare variants [< 1% in public databases, including 1000 Genomes project, NHLBI Exome Variant Server, Complete Genomics 69, and Exome Aggregation Consortium (ExAC v0.2)] that were fitting a recessive or a de novo model. After identifying the UGP2 variant in the proband, Sanger sequencing was used to confirm segregation in other affected and unaffected family members.
Individuals 11, 20, 21 and 22
Whole exome sequencing was performed at CENTOGENE AG, as previously described [105].
Individuals 12 and 13
High-quality DNA was used to capture exons using the SureSelect kit (Agilent, Santa Clara, CA, US). Then genomic libraries were created according to the manufacturer’s protocols. Sequences were read on Proton (Life Technologies Inc., Carlsbad, CA, US). Downstream analyses such as sequence alignment, indexing and raw variant calling were done using publicly and commercially available tools such as Ion Reporter, SAMTools, and Genomic Analysis ToolKit. Moreover, variant interrogations were done using sequence-variant databases, such as dbSNP, Ensembl, and the National Heart, Lung, and Blood Institute (NHLBI) Exome Variant Server (EVS), 1000 genome project.
Individual 19
Whole exome sequencing was performed in a diagnostic setting at MEDGENOME, India. DNA extracted from blood was used to perform targeted gene capture using the Agilent SureSelect V5 exome capture kit. The libraries were sequenced to mean > 80–100 × coverage on Illumina sequencing platform. GATK best practices framework was used for variant identification using Sentieon (v201808.01), sequences obtained were aligned to GRCh37/hg19 using Sentieon aligner and analyzed using Sentieon for removing duplicates, recalibration and re-alignment of indels. Sentieon haplotypecaller has been used to identify variants which are relevant to the clinical indication. Gene annotation of the variants was performed using VEP program against the Ensemble release 91 human gene model.
Human brain samples
Tissue was obtained, upon informed consent, and used in a manner compliant with the Declaration of Helsinki and the Research Code provided by the local ethical committees. Fetal brains were preserved after spontaneous or induced abortions with appropriate written consent for brain autopsy and use of rest material for research. We performed a careful histological and immunohistochemical analysis, and evaluation of clinical data (including genetic data, when available). We only included specimens displaying a normal cortical structure for the corresponding age and without any significant brain pathology.
Brain tissue immunohistochemistry
For immunohistochemical analysis, we used two cases from the first trimester (GW6 and GW9), four cases from the second trimester (GW21, GW23, GW24 and GW26) and two cases from the third trimester (GW33 and GW36). Anatomical regions were determined according to the atlas of human brain development [11,12,13,14]. We cut 4-µm sections from formalin-fixed, paraffin-embedded whole fetuses (GW6 and GW9) and brain tissue from cerebral, mesencephalic, cerebellar and brain stem regions (from GW21 to GW36). Slides were stained with mouse anti-UGP2 (C-6) in a 1:150 dilution (Santa Cruz) and visualized using Mouse and Rabbit Specific HRP/DAB (ABC) Detection IHC kit (Abcam). Mayer’s hematoxylin was used as a counterstain for immunohistochemistry followed by mounting and coverslipping (Bio-Optica) for slides. Prepared slides were analyzed and scanned under a VisionTek® Live Digital Microscope (Sakura).
Cloning of UGP2 cDNA
RNA was isolated using TRI reagent (Sigma) from whole peripheral blood of index patient 1 and her parents, after red blood cell depletion with RBC lysis buffer (168 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA). cDNA was synthesized following the iSCRIPT cDNA Synthesis Kit (Bio-Rad) protocol, and the coding sequence of the long and short UGP2 isoform (wild type or mutant) was PCR amplified together with homology arms for Gibson assembly (see Supplementary Table 8, online resource, for primer sequences) using Phusion High-Fidelity DNA polymerase (NEB). PCR-amplified DNA was then cloned by Gibson assembly as previously described [9] in a pPyCAG-IRES-puro plasmid (a kind gift from Ian Chambers, Edinburgh) opened with EcoRI for experiments in mammalian cells. All obtained plasmids were sequence verified by Sanger sequencing (complete plasmid sequences available upon request).
Fibroblast cell culture
Fibroblasts from index patient 1 and her parents were obtained using a punch biopsy according to standard procedures, upon informed consent (IRB approval MEC-2017-341). Fibroblasts from the parents of index patients 2 and 3 were also obtained upon informed consent at McMaster Children’s Hospital. All fibroblasts were cultured in standard DMEM medium supplemented with 15% fetal calf serum, MEM non-essential amino acids (Sigma), 100 U/ml penicillin and 100 µg/ml streptomycin, as done previously [6], in routine humidified cell culture incubators at 20% O2. Fibroblast cell lines were transfected using Lipofectamine 3000 (Invitrogen) with the indicated plasmid constructs. All cell lines used in this report were regularly checked for the presence of mycoplasma and were negative during all experiments.
Genome engineering in human embryonic stem cells
H9 human embryonic stem cells were cultured as previously described [8, 9]. In short, cells were maintained on feeder-free conditions in mTeSR-1 medium (STEMCELL technologies) on Matrigel (Corning)-coated culture dishes. To engineer the patient-specific UGP2 mutation by homologous recombination [7], ESCs were transfected using Lipofectamine 3000 with a plasmid expressing eSpCas9-t2a-GFP (a kind gift of Feng Zhang) and a gRNA targeting the UGP2 gene (see Supplementary Table 8, online resource, for the sequence), together with a 60-bp single-stranded oligonucleotide (ssODN) homology template encoding the patient mutation (synthesized at IDT). To increase the stability of the ssODN and, therefore, homologous recombination efficiency, the first two 5′ and 3′ nucleotides were synthesized using phosphorothiorate bonds [80]. 48 h post-transfection, GFP-expressing cells were sorted, and 6000 single GFP-positive cells were plated on a Matrigel-coated six-well plate in the presence of 10 µM ROCK-inhibitor (Y27632, Millipore). After approximately 10 days, single colonies where manually picked, expanded and genotyped using Sanger sequencing (see Supplementary Table 8, online resource, for primer sequences). As a by-product of non-homologous end joining, knockout clones were obtained which showed a single nucleotide A insertion at position 42 of UGP2 transcript 1 (chr2:64083462_64083463insA), leading to an out-of-frame transcript and a premature termination of the protein at amino acid position 47 (D15Rfs*33). Western blotting confirmed the absence of all UGP2 proteins in knockout clones and the loss of the short UGP2 isoform in clones with the patient mutation. To produce a stable rescue cell line, ESCs were transfected as previously described with the pPyCAG-IRES-puro plasmid expressing either the long WT or mutant UGP2 isoform. After 48 h, the population of cells with the transgene integration was selected with 1 µg/ml puromycin. Engineered ESC clones had a normal colony morphology and pluripotency factor expression.
Patient-specific induced pluripotent stem cell generation
Patient fibroblast cell lines were reprogrammed using the CytoTune™-iPS 2.0 Sendai Reprogramming Kit (Thermo Scientific, A16517) expressing the reprogramming factors OCT4, SOX2, KLF4 and C-MYC on Matrigel-coated cell culture plates, upon informed consent (IRB approval MEC-2017–341). After approximately 4–5 weeks, emerging colonies were manually picked and expanded. Multiple clones were assessed for their karyotype, pluripotency factor expression and three lineage differentiation potential (Stem Cell Technologies, #05230), following the routine procedures of the Erasmus MC iPS Cell core facility, as previously described [6]. Sanger sequencing was used to verify the genotype of each obtained iPSC line. We used three validated clones for each individual in our experiments.
Neural stem cell differentiation
Pluripotent cells were differentiated in neural stem cells (NSCs), using a modified dual SMAD inhibition protocol [20]. In short, 18,000 cells/cm2 were plated on Matrigel-coated cell culture dishes in mTeSR-1 medium in the presence of 10 µM Y27632. When cells reached 90% confluency, the medium was switched to differentiation medium (KnockOut DMEM (Gibco), 15% KnockOut serum replacement (Gibco), 2 mM l-glutamine (Gibco), MEM non-essential amino acids (Sigma), 0.1 mM β-mercaptoethanol, 100U/ml penicillin and 100 µg/ml streptomycin) supplemented with 2 µM A 83-01 (Tocris) and 2 µM Dorsomorphin (Sigma-Aldrich). At day 6, medium was changed to an equal ratio of differentiation medium and NSC medium (KnockOut DMEM-F12 (Gibco), 2 mM l-glutamine (Gibco), 20 ng/ml bFGF (Peprotech), 20 ng/ml EGF (Peprotech), 2% StemPro Neural supplement (Gibco), 100U/ml penicillin and 100 µg/ml streptomycin) supplemented with 2 µM A 83-01 (Tocris) and 2 µM Dorsomorphin (Sigma-Aldrich). At day 10, cells were passaged (NSC p = 0) using Accutase (Sigma) and maintained in NSC medium. We used commercially available H9-derived NSCs (Gibco) as a control (a kind gift from Raymond Poot, Rotterdam).
Other stem cell differentiation experiments
ESCs were differentiated into hematopoietic stem cells and cardiomyocytes using commercially available STEMCELL technology kits (STEMdiff Hematopoietic kit #05310, STEMdiff Cardiomyocyte differentiation kit #05010) according to the manufacturer’s instructions. Cells were finally harvested and lysed with TRI reagent to isolate RNA for further qRT-PCR analysis.
RNA-sequencing and data analysis
For patient RNA-seq, peripheral blood was obtained from index patient 1 and her parents, collected in PAX tubes and RNA was isolated following standard diagnostic procedures in the diagnostics unit of the Erasmus MC Clinical Genetics department. RNA-seq occurred in a diagnostic setting, and sequencing was performed at GenomeScan (Leiden, The Netherlands). For RNA-seq of in vitro-cultured cell lines, RNA was obtained from six-well cultures using TRI reagent, and further purified using column purification (Qiagen, #74204). mRNA capture, library prep including barcoding and sequencing on an Illumina HiSeq2500 machine were performed according to standard procedures of the Erasmus MC Biomics facility. Approximately 20 million reads were obtained per sample. For cell line experiments, two independent H9 wild-type cultures, two independent knockout clones harboring the same homozygous UGP2 genetic alteration and two independent clones harboring the patient homozygous UGP2 mutation were used. Each cell line was sequenced in two technical replicates at ESC state and differentiated NSC state (at passage 5). FASTQ files obtained after de-multiplexing of single-end, 50-bp sequencing reads were trimmed by removing possible adapters using Cutadapt after quality control checks on raw data using the FastQC tool. Trimmed reads were aligned to the human genome (hg38) using the HISAT2 aligner [50]. To produce Genome Browser Tracks, aligned reads were converted to bedgraph using bedtools genomecov, after which the bedGraphToBigWig tool from the UCSC Genome Browser was used to create a bigwig file. Aligned reads were counted for each gene using htseq-count [3] and GenomicFeatures [55] was used to determine the gene length by merging all non-overlapping exons per gene from the Homo_sapiens.GRCh38.92.gtf file (Ensemble). Differential gene expression and RPKM (Reads Per Kilobase per Million) values were calculated using edgeR [85] after removing low-expressed genes and normalizing data. The threshold for significant differences in gene expression was FDR < 0.05. To obtain a list of ESC and NSC reference genes used in Supplementary Fig. 6F, online resource, we retrieved genes annotated in the following GO terms using GSEA/MSigDB web site v7.0: GO_FOREBRAIN_NEURON_DEVELOPMENT (GO:0021884), GO_CEREBRAL_CORTEX_DEVELOPMENT (GO:0021987), GO_NEURAL_TUBE_DEVELOPMENT (GO:0021915), BHATTACHARYA_EMBRYONIC_STEM_CELL (PMID: 15070671) and BENPORATH_NOS_TARGETS (PMID: 18443585).
Functional enrichment analysis
Metascape [123], g:profiler [79] and Enrichr [52] were used to assess functional enrichment of differentially expressed genes. Supplementary Table 4, online resource, reports all outputs in LogP, log(q value) and Adjusted p value (q value) for Metascape and g:profiler, and in p value, Adjusted p value (q value) and combined score (which is the estimation of significance based on the combination of Fisher's exact test p value and z score deviation from the expected rank) for Enrichr. All tools were used with default parameters and whole genome set as background.
Genome-wide homology search
To make a genome-wide list of transcripts sharing a similar structure as UGP2 transcripts, 42,976 transcripts from 21,522 genes (Human genes GRCh38.p12) were extracted using BioMart of Ensembl (biomaRt R package). 11,056 out of 21,522 genes had only 1 transcript and the remaining 31,920 transcripts from 10,466 genes were selected, the protein sequences were obtained with biomaRt R package and homology analysis was performed using the NCBI’s blastp (formatting option: -outfmt = 6) command line. We grouped longest and shorter transcript based on coding sequence length and only kept those that matched a pairwise homology comparison between the longest and the shorter transcript with the following criteria: complete 100 percent identity, without any gap and mismatch, and starting ATG codon of shortest transcript being part of the longest transcript(s). 1766 genes meet these criteria. We then filtered these genes for published essential genes [10], leaving us with 1197 genes. Using BioMart (Attributes: Phenotype description and Study external reference) of Ensembl we then evaluated the probability that these genes were implicated in disease and identified 850 genes that did not have an association with disease phenotype/OMIM number. Of those, 247 genes encoded proteins of which the shorter isoform differed less than 50 amino acids from the longer isoform. We chose this arbitrary threshold to exclude those genes where both isoforms could encode proteins differing largely in size and might, therefore, encode functionally completely differing proteins (although we cannot exclude that this will also hold true for some of the genes in our selection).
Differential isoform expression in fetal tissues
Publically available RNA-seq data from various fetal tissue samples (Supplementary Table 2, online resource) were analyzed using the same workflow as described for the RNA-seq data analysis above. To determine differential isoform expression in these tissues, we calculated a ratio between the unique exon(s) of the shortest and longest transcript for each gene and assessed its variability across different fetal tissue samples. The number of reads for each unique exon of a transcript was calculated by mapping aligned RNA-seq reads against the unique exon coordinate using bedtools multicov. The longest and shortest transcripts were separated and the transcript ratio (number of counts of shortest transcript/(number of counts of shortest transcript + number of counts of longest transcript)) for each gene was obtained from the average reads of RNA-seq samples per tissue. 382 genes out of 1197 genes showed high variability across different samples (defined as a difference between highest and lowest ratio > 0.5), 277 of those highly variable genes were not associated with a disease phenotype/OMIM number and of these 83 genes had a length less than 50 amino acids (a subset of the 247 genes with no OMIM and length less than 50 amino acids).
Haplotype analysis
The 30 MB region surrounding UGP2 was extracted from exome sequencing VCF files to include both common and rare polymorphisms. Variants were filtered for a minimum depth of coverage of at least 10 reads and a genotype quality of at least 50. The filtered variants were then used as input in PLINK (v1.07) with the following settings:
-
homozyg-snp 5
-
homozyg-kb 100
-
homozyg-gap 10,000
-
homozyg-window-het 0
ROH around the UGP2 variant was identified in all five probands examined. The minimum ROH in common between all samples was a 5-Mb region at chr2: 60679942–65667235. We note that targeted sequencing leads to uneven SNP density, so the shared ROH may, in fact, be larger or smaller. Next, we used recombination maps from deCODE to estimate the size of the region in centiMorgans (cM). We then used the region size in cM to estimate the time to event in generations using methods previously described [120].
qPCR analysis
RNA was obtained using TRI reagent, and cDNA prepared using iSCRIPT cDNA Synthesis Kit according to the manufacturer’s instructions. qPCR was performed using iTaq universal SYBR Green Supermix in a CFX96RTS thermal cycler (Bio-Rad). Supplementary Table 8, online resource, summarizes all primers used in this study. Relative gene expression was determined following the ΔΔct method. To calculate the ratio of the short isoform, we performed absolute quantification as previously described [109]. Briefly, we performed qPCR on known copy numbers, ranging from 103 to 108 copies, of a plasmid containing the short UGP2 isoform (5′ UTR included) using primers detecting specifically either the total or the short isoform. After plotting the log copy number versus the ct, we obtained a standard curve that we used to extrapolate the copy number of the unknown samples. To test for significance, we used Student’s T test and considered p < 0.05 as significant.
Western blotting
Proteins were extracted with NE buffer (20 mM HEPES, pH 7.6, 1.5 mM MgCl2, 350 mM KCl, 0.2 mM EDTA and 20% glycerol) supplemented with 0.5% NP40, 0.5 mM DTT, cOmplete Protease Inhibitor Cocktail (Roche) and 150 U/ml benzonase. Protein concentration was determined by BCA (Pierce) and 20–50 µg of proteins was loaded onto a 4–15% Criterion TGX gel (Bio-Rad). Proteins were then transferred to a nitrocellulose membrane using the Trans-Blot Turbo Transfer System (Bio-Rad). The membrane was blocked in 5% milk in PBST and subsequently incubated overnight at 4 °C with primary antibody diluted in milk. After PBST washes, the membrane was incubated 1 h at RT with the secondary antibody and imaged with an Odyssey CLX scanning system (Li-Cor). Band intensities were quantified using Image Studio (Li-cor). Antibodies used were Ms-α-UGP2 (sc-514174) 1:250; Ms-α-Vinculin (sc-59803) 1:10,000; Gt-α-actin (sc-1616) 1:500; Ms-α-LAMP2 (H4B4) 1:200; IRDye 800CW Goat anti-Mouse (926-32210) 1:5000; IRDye 680 Donkey anti-Goat (926-32224) 1:5000.
Zebrafish disease modeling
Animal experiments were approved by the Animal Experimentation Committee at Erasmus MC, Rotterdam. Zebrafish embryos and larvae were kept at 28 °C on a 14–10‐h light–dark cycle in 1 M HEPES buffered (pH 7.2) E3 medium (34.8 g NaCl, 1.6 g KCl, 5.8 g CaCl2·2H2O, 9.78 g MgCl2·6H2O). For live imaging, the medium was changed at 1 dpf to E3 + 0.003% 1‐phenyl 2‐thiourea (PTU) to prevent pigmentation. Ugp2a and ugp2b were targeted by Cas9/gRNA RNP complex as we did before [51]. Briefly, fertilized oocytes from a tgBAC(slc1a2b:Citrine)re01tg reporter line [51] maintained on an TL background strain were obtained, and injected with Cas9 protein and crRNA and tracrRNA synthesized by IDT (Alt-R CRISPR–Cas9 System), targeting the open reading frame of zebrafish ugp2a and ugp2b. DNA was extracted from fin clips and used for genotyping using primers flanking the gRNA location (Supplementary Table 8, online resource) followed by sequencing. Mutants with a high level of out-of-frame indels in both genes were identified using TIDE [18] and intercrossed to obtain germ line transmission. Upon re-genotyping, mutant zebrafish with the following mutations as indicated in Fig. 6 were selected and further intercrossed. In this study, we describe two new mutant fish lines containing deletions in ugp2a (ugp2aΔ/Δ) and ugp2b (ugp2bΔ/Δ): ugp2are08/re08 containing a 37 bp deletion in exon 2 and ugp2bre09/re09 containing a 5 bp deletion in exon 2. Intravital imaging, and analysis of eye movement, was performed as previously described [51]. Briefly, zebrafish larvae anesthetized in tricaine were mounted in low-melting point agarose-containing tricaine and imaged using a Leica SP5 intravital imaging setup with a 20 × /1.0 NA water-dipping lens. To assess the locomotor activity of zebrafish larvae from 3 to 5 dpf, locomotor activity assays were performed using an infrared camera system (DanioVision™ Observation chamber, Noldus) and using EthoVision® XT software (Noldus) as described [51]. Briefly, control (n = 24) and ugp2aΔ/Δ; ugp2bΔ/Δ (n = 24) zebrafish larvae, in 48-well plates, were subjected to gradually increasing (to bright light) and decreasing light conditions (darkness) as in Kuil et al. [51]. Distance traveled (mm) per second was measured. For 4-AP (Sigma) stimulation, animals were treated with 4-AP dissolved in DMSO 30 min before the onset of the experiments. For these experiments, locomotor activity was measured over 35 min, with the first 5 min going from dark to light, followed by 30 min under constant light exposure.
Periodic acid–Schiff (PAS) staining
ESCs or differentiated NSCs (wild type, KO, KI or rescue) were incubated under hypoxia conditions (3% O2) for 48 h. Cells were fixed with 5.2% formaldehyde in ethanol, incubated 10 min with 1% periodic acid, 15 min at 37 °C with Schiff’s reagent (Merck) and 5 min with hematoxylin solution (Klinipath) prior to air drying and mounting. Every step of the protocol is followed by a 10-min wash with tap water. Imaging occurred on an Olympus BX40 microscope. Images were acquired at a 100 × magnification, and ImageJ software was used for quantification. For ESCs, we used a minimum of 20 images per genotype for the quantification, containing on average 20 cells each, calculating the percentage of PAS-positive area. For NSCs, we imaged between 80 and 100 cells per genotype, counting the number of glycogen granules in the cytoplasm. We report the average of two independent experiments at 48 h low oxygen.
UGP2 enzymatic activity
The measurement of UGP2 enzyme activity was performed according to a modified GALT enzyme activity assay as described previously [62]. Frozen cell pellets were defrosted and homogenized on ice. 10 µl of each cell homogenate (around 0.5 mg protein/ml as established by BSA protein concentration determination) was pre-incubated with 10 µl of dithiothreitol (DDT) for 5 min at 25 °C. 80 µl of a mixture of glucose-1-phosphate (final concentration 1 mM), UTP (0.2 mM), magnesium chloride (1 mM), glycine (125 mM) and Tris–HCl (pH8) (40 mM) was added and incubated for another 15 min at 25 °C. The reaction was stopped by adding 150 µl of 3.3% perchloric acid. After 10 min on ice, the mixture was centrifuged (10,000 rpm for 5 min at 4 °C), the supernatant isolated and neutralized with ice-cold 8 µl potassium carbonate for 10 min on ice. After centrifugation, the supernatant was isolated and 1:1 diluted with eluent B (see below) after which the mixture was added to a MilliPore Amicon centrifugal filter unit. After centrifugation, the supernatant was stored at − 20 °C until use. The separation was performed by injection of 10 µl of the defrosted supernatant onto a HPLC system with UV/VIS detector (wavelength 262 nm) equipped with a reversed-phase Supelcosil LC-18-S 150 mm × 4.6 mm, particle size 5 µm, analytical column and Supelguard LC18S guard column (Sigma-Aldrich). During the experiments, the temperature of the column was maintained at 25 °C. The mobile phase consisted of eluent A (100% methanol) and eluent B (50 mM ammonium phosphate buffer pH 7.0 and 4 mM tetrabutylammonium bisulphate). A gradient of 99% eluent B (0–20 min), 75% eluent B (20–30 min) and 99% eluent B (30–45 min) at a flow rate of 0.5 m/min was used. The reaction product UDP-glucose was quantified using a calibration curve with known concentrations of UDP-glucose. UGP2 activity was expressed as the amount of UDP-glucose formed per mg protein per min. Experiments were performed in duplicate and for every cell line two independently grown cell pellets were used.
Immunostaining/immunohistochemistry
For immunofluorescence staining, cells were seeded on coverslips coated with 100 µg/ml poly-d-lysine (Sigma) overnight. For ESC, coverslips were further coated with Matrigel (Corning) for 1 h at 37 °C. At 70% confluency, cells were fixed with 4% PFA for 15 min at RT. Cells were then permeabilized with 0.5% Triton in PBS, incubated 1 h in blocking solution (3% BSA in PBS) and then overnight at 4 °C with the primary antibody diluted in blocking solution. The next day coverslips were incubated 1 h at room temperature in the dark with a Cy3-conjugated secondary antibody and mounted using ProLong Gold antifade reagent with DAPI (Invitrogen) to counterstain the nuclei. Images were acquired with a ZEISS Axio Imager M2 using a 63X objective.
Data availability
RNA-Seq of in vitro studies is publicly available through the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) under accession number GSE137129. Due to privacy regulations and consent, raw RNA-seq data from patient blood and genomic sequencing data cannot be made available. To retrieve tissue wide expression levels of UGP2, the GTEx Portal was accessed on 16/07/2019 (https://gtexportal.org/home/). RNA-seq data from various tissues were downloaded from various publications [46, 83, 94, 118]. All publically available data that were re-analyzed here are summarized in Supplementary Table 2, online resource.
Results
A recurrent ATG mutation in UGP2 in 22 individuals presenting with a severe DEE
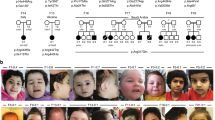
We encountered a 3-month-old girl (Fig. 1a, family 1, individual 1) that was born as the first child to healthy non-consanguineous Dutch parents, by normal vaginal delivery after an uneventful pregnancy conceived by ICSI. She presented in the first weeks of life with irritability and jitteriness, which developed into infantile spasms and severe epileptic activity on multiple electroencephalograms, giving rise to a clinical diagnosis of West syndrome (Fig. 1b). Despite the use of multiple anti-epileptic drugs, including ACTH and a ketogenic diet, seizures remained intractable and occurred daily. Severe developmental delay was evident without acquisition of any noticeable developmental milestones, causing the need for gastrointestinal tube feeding. Visual tracking was absent, and foveal hypopigmentation, hypermetropia and mild nystagmus were noticed upon ophthalmological investigation. MRI brain imaging showed no gross structural abnormalities or migration disorders at the age of 4 months, but displayed reduced white matter, that further developed into global atrophy with wide sulci and wide pericerebral liquor spaces at the age of 17 months (Fig. 1c, Supplementary Fig. 1b, online resource). At that time, she had become progressively microcephalic, with a head circumference of − 2.96 SD at the last investigation at 23 months of age (Supplementary Fig. 1a, online resource). She showed a number of minor dysmorphisms, including a sloping forehead, elongated head with suture ridging, bitemporal narrowing, a relatively small mouth and large ears (Fig. 1a). Neurological examination showed brisk, symmetric deep tendon reflexes, more pronounced at the upper limbs. Routine investigations, including metabolic screening in urine, plasma and cerebrospinal fluid were normal. A SNP-array showed a normal female chromosomal profile, with a large, ~ 30 Mb run of homozygosity (ROH) at chromosome 2, and a few smaller ROH regions, adding up to 50 Mb ROH regions in total, pointing to an unrecognized common ancestor of both parents (coefficient of inbreeding 1/64). Subsequent trio WES did not show any disease-causing variants in known DEE genes, but identified a homozygous variant (chr2:64083454A>G) in UGP2, located in the large ROH region (Fig. 1d), with no other disease-implicated variants observed in that region. Both parents were heterozygous carriers of the same variant. Via Genematcher [97] and our network of collaborators, we identified 21 additional individuals from 14 unrelated families (of which 10 were consanguineous), harboring the exact same homozygous variant and presenting with an almost identical clinical phenotype of intractable seizures, severe developmental delay, visual disturbance, microcephaly and similar minor dysmorphisms (Fig. 1a, c, e, Supplementary Fig. 1b, Supplementary Case reports, Supplementary Movie 1, Supplementary Table 1, online resource, for detailed information on 18 cases). Ten of these individuals passed away early, with the majority before the age of 3.5 years. In six families, at least seven already deceased siblings had a similar phenotype but could not be investigated. Two families were of Indian descent (both with ancestors from regions currently belonging to Pakistan), living in Canada (family 2) and the USA (family 3), with the remaining families from Oman (family 4, originally from Pakistan), Pakistan (family 5, family 13), Iran (families 6, 7, 8 and 11), UAE (family 9), Saudi Arabia (family 10) and India (family 12). Two additional cases in family 14 from Oman and family 15 from India were identified presenting with intractable seizures and microcephaly, but no detailed medical information could be obtained at this point.
UGP2 homozygous variants in 20 individuals with severe epileptic encephalopathy. a Facial pictures of individual 1 (at 18 and 23 months), individual 5 (at 9 years), individual 6 (at 11 months), individual 9 (at 18 months), individual 10 (at 2 years) and individual 19 (at 13 months). Note the progressive microcephaly with sloping forehead, suture ridging, bitemporal narrowing, high hairline, arched eyebrows, pronounced philtrum, a relatively small mouth and large ears. b Electroencephalogram of individual 1 at the age of 8 months showing a highly disorganized pattern with high-voltage irregular slow waves intermixed with multifocal spikes and polyspikes. c T1-weighted mid-sagittal brain MRI of individual 1 (at 17 months) and individual 4 (at 24 months) illustrating global atrophy and microcephaly but no major structural anomalies. d Sanger sequencing traces of family 1, confirming the chr2:64083454A>G variant in UGP2 in heterozygous and homozygous states in parents and affected individual 1, respectively. e Family pedigrees of ascertained patients. Affected individuals and heterozygous parents are indicated in black and half black, respectively. Affected individuals with confirmed genotype are indicated with an arrow, and numbers. Other not-tested affected siblings presenting with similar phenotypes are indicated with a question mark. Consanguineous parents are indicated with a double connection line. Males are squares, females are circles; unknown sex is indicated with rotated squares; deceased individuals are marked with a line. f Violin plots showing distribution of gene expression (in TPM) amongst male and female samples from the GTEx portal for various brain regions. Outliers are indicated by dots. g Multiple species sequence alignment from the UCSC browser, showing that the ATG start site is highly conserved
Having identified at least 22 individuals with an almost identical clinical phenotype and an identical homozygous variant in the same gene led us to pursue UGP2 as a candidate gene for a new genetic form of DEE. UGP2 is highly expressed in various brain regions (Fig. 1f), and also widely expressed amongst other tissues, including liver and muscle according to the data from the GTEx portal [23] (Supplementary Fig. 1d, online resource). The (chr2:64083454A>G) variant is predicted to cause a missense variant (c.34A>G, p.Met12Val) in UGP2 isoform 1 (NM_006759), and to cause a translation start loss (c.1A>G, p.?.) of UGP2 isoform 2 (NM_001001521), referred to as long and short isoforms, respectively. The variant has not been reported in the Epi25 web browser [31], ClinVar [54], LOVD [37], Exome Variant Server [33], DECIPHER [35], GENESIS [40], GME variome [90] or Iranome databases [34], is absent from our in-house data bases and is found only 15 times in a heterozygous, but not homozygous, state in the 280,902 alleles present in gnomAD (MAF: 0.00005340) [56]. In the GeneDx unaffected adult cohort, the variant was found heterozygous 10 times out of 173,502 alleles (MAF: 0.00005764), in the ~ 10,000 exomes of the Queen Square Genomic Center database two heterozygous individuals were identified, and out of 45,921 individuals in the Centogene cohort, 10 individuals are heterozygous for this variant. The identified variant has a CADD score (v1.4) of 19.22 [81] and Mutation Taster [89] predicted this variant as disease causing. The nucleotide is strongly conserved over multiple species (Fig. 1 g). Analysis of WES data from 5 patients did provide evidence of a shared ROH between patients from different families (including the Dutch family), indicating that this same variant might represent an ancient mutation that originated some 26 generations ago (Supplementary Fig. 1c, online resource). Interestingly, since most families originally came from regions of India, Pakistan and Iran, overlapping with an area called Balochistan, this could indicate that the mutation has originated there around 600 years ago. As Dutch traders settled in that area in the seventeenth century, it is tempting to speculate that this could explain the co-occurrence of the variant in these distant places [1].
Short UGP2 isoform is predominantly expressed in brain and absent in patients with ATG mutations
Both UGP2 isoforms only differ by 11 amino acids at the N-terminal (Fig. 2a) and are expected to be functionally equivalent [38]. To investigate how the A>G variant may cause DEE, we first obtained fibroblasts from individual 1 (homozygous for the A>G variant) and her heterozygous parents and analyzed the isoform expression by Western blotting (Fig. 2b). Whereas the two isoforms were equally expressed in wild-type fibroblasts, the expression of the shorter isoform was diminished to ~ 25% of total UGP2 in heterozygous parents, both of individual 1 (Fig. 2b, c) and of individuals 2 and 3 (Supplementary Fig. 2a, b, online resource), and was absent in cells from the affected individual 1 (Fig. 2b, c; fibroblasts of the affected children in family 2 or other families were not available). Total UGP2 levels were not significantly different between the affected child and her parents, or between parents and wild-type controls (Fig. 2d, Supplementary Fig. 2c, online resource). This indicates that the long isoform harboring the Met12Val missense variant is upregulated in fibroblast when the short isoform is missing. Moreover, this indicates that Met12Val does not affect the stability of the long isoform at the protein or transcript level (Supplementary Fig. 2d–f, online resource). RNA-seq on peripheral blood samples of family 1 did not identify altered splicing events of UGP2 and the global transcriptome of the proband was not different from her parents, although only a limited analysis could be performed as only a single sample was available for each individual (Supplementary Fig. 2g, h, online resource). Both homozygous and heterozygous fibroblasts had a similar proliferation rate compared to wild-type fibroblasts (Fig. 2e, Supplementary Fig. 2i, online resource), and immunocytochemistry confirmed a similar subcellular localization of UGP2 in mutant and wild-type cells (Fig. 2f). We then measured the enzymatic activity of UGP2 in wild type, heterozygous and homozygous fibroblasts, and found that mutant fibroblast had a similar capacity to produce UDP-glucose in the presence of exogenously supplied glucose-1-phosphate and UTP (Fig. 2g). Altogether, this indicates that the long UGP2 isoform harboring the Met12Val missense change is functional and is, therefore, unlikely to contribute to the patient phenotype.
UGP2 homozygous variant leads to a loss of the shorter protein isoform in patient fibroblasts. a Schematic drawing of the human UGP2 locus, with both long and short transcript isoforms. Boxes represent exons, with coding sequences indicated in green. The location of the recurrent mutation is indicated in both transcripts. b Western blotting of cellular extracts derived from control fibroblasts and fibroblasts obtained from family 1, detecting the housekeeping control vinculin and UGP2. Note the two separated isoforms of UGP2 that have a similar intensity in wild-type cells. The shorter isoform is less expressed in fibroblasts from heterozygous parents and absent in fibroblasts from the affected proband. c Western blot quantification of the fraction of short UGP2 protein isoform compared to total UGP2 expression in control, parental heterozygous and proband homozygous fibroblasts, as determined in three independent experiments. Error bars represent SEM. d Western blot quantification of total UGP2 protein levels, as determined by the relative expression to the housekeeping control vinculin. Bar plot showing the results from three independent experiments. Error bars represent SEM; no significant differences were found between parents and proband, t test, two tailed. e Cell proliferation experiment of fibroblasts from heterozygous parents and homozygous proband from family 1, during a 5-day period, determined in three independent experiments. Error bars represent SEM. f Immunocytochemistry on cultured control and UGP2 heterozygous and homozygous mutant fibroblasts derived from family 1, detecting UGP2 (red). Nuclei are stained with DAPI. Scale bar 50 µm. g Enzymatic activity of UGP2 in control and UGP2 heterozygous and homozygous mutant fibroblasts derived from family 1. Shown is the mean of two independent experiments. Error bars represent SEM; no significant differences were found, unpaired t test, two tailed
As the A>G variant results in a functional long UGP2 isoform but abolishes the translation of the shorter UGP2 isoform, we next investigated whether the ratio between short and long isoform differs amongst tissues. If so, the homozygous A>G variant would lead to depletion of UGP2 in tissues where mainly the short isoform is expressed, possibly below a threshold that is required for normal development or function. Western blotting on cellular extracts derived from wild-type H9 human embryonic stem cells (ESCs), commercially acquired H9-derived neural stem cells (NSCs) and fibroblasts (Fig. 3a) showed that, whereas the ratio between short and long isoform in fibroblasts was around 0.5, in ESCs it was 0.14 and in NSCs 0.77, indicating that the shorter UGP2 isoform is the predominant one in NSCs (Fig. 3b). A similar trend was observed when assessing the transcript level, both by multiplex RT-PCR and qRT-PCR, using primers detecting specifically the short and long transcript isoform (Fig. 3c–e). This indicates that differential isoform expression between cell types is regulated at the transcriptional level, possibly hinting at tissue-specific regulatory elements driving isoform expression. We next analyzed RNA-seq data from human fetal tissues [46, 83, 94, 118] to determine the fraction of reads covering short versus total UGP2 transcripts (Fig. 3f). This showed that in human fetal brain the short transcript isoform is predominantly expressed. To gain more insight into the cell type-specific expression of UGP2, we performed immunohistochemistry on human fetal brain tissues from the first to third trimester of pregnancy (Fig. 3g). In the first trimester, we found pale labeling of neuropil in the proliferative neuroepithelium of the hypothalamic, cortical, mesencephalic and thalamic regions (Fig. 3g-A/I, II, III, IV), as well as the marginal zone of the spinal cord (Fig. 3g-A/V) and cuboidal epithelial cells of choroid plexus (Fig. 3g-A/VI). During the second trimester, UGP2 positivity was detected in neurons from the subplate region of the cerebral cortex (Fig. 3g-B/I, II) and still in some of the cells in the neuroepithelium and subventricular zone (Fig. 3g-B/III). Almost the same pattern of UGP2 distribution was found in the cerebral cortex of fetuses from the third trimester. Also, we found clear cytoplasmatic UGP2 expression in neurons from mesencephalic, inferior olivary and cerebellar nuclei during the second (Fig. 3g-B/IV, V, and VI) and third trimester, respectively (Fig. 3g-C/IV, V). In the white matter of the cerebellum in the third trimester, we identified single-positive glial cells (Fig. 3g-C/VI). In the cerebellar cortex, we did not find specific positivity of cells on UGP2 (Fig. 3g-B, C/VII). Cuboidal epithelial cells of choroid plexus preserved UGP2 positivity during the second trimester (Fig. 3g-B/VIII) but lost it in the third trimester (Fig. 3g-C/VIII). Together this indicates that UGP2 can be detected in a broad variety of cell types during brain development. On Western blotting, we noticed preferential expression of the shorter UGP2 isoform in the developing cortex and cerebellum from gestational weeks 14, 20 and 28 (Fig. 3h) and in the frontal cortex of brains from weeks 21 and 23 (Supplementary Fig. 2j, online resource). Together, this supports the hypothesis that the DEE phenotype in patients is caused by a major loss of functional UGP2 in the brain, as the short isoform represents virtually all UGP2 produced in this tissue.
UGP2 short isoform is predominant in brain-related cell types. a Western blotting showing UGP2 expression in H9 human embryonic stem cells (ESCs), H9-derived neural stem cells (NSCs) and fibroblasts (Fibro). Vinculin is used as a housekeeping control. Note the changes in relative expression between the two UGP2 isoforms in the different cell types. L, ladder. b Western blot quantification of the fraction of short UGP2 protein isoform compared to total UGP2 expression, as determined in three independent experiments. Error bars represent SEM. c Multiplex RT-PCR of ESCs, NSCs and fibroblasts, showing a similar variability in isoform expression at the transcript and at the protein level. Each cell line was tested in triplicates. d Quantification of the fraction of the short UGP2 transcript isoform compared to total UGP2 expression, from the multiplex RT-PCR from c. Error bars represent SEM. e Quantification of the fraction of short UGP2 transcript isoform compared to total UGP2 expression by qRT-PCR in three independent experiments. Error bars represent SEM. f Ratio of RNA-seq reads covering the short transcript isoform compared to the total reads (covering both short and long isoforms), in multiple fetal tissues. In RNA-seq samples derived from brain, virtually all UGP2 expressions come from the short isoform. Error bars represent SD. g Immunohistochemistry detecting UGP2 in human fetal brains from the first, second and third trimester (gestational week (GW) 6, 9, 23 and 36). See text for details. h Western blotting detecting UGP2 in various human brain regions at weeks 14, 20 and 28 of gestation, showing the virtual absence of the long isoform expression in fetal brain. Vinculin is used as a housekeeping control. L ladder
Lack of the short UGP2 isoform leads to transcriptome changes upon differentiation into neural stem cells
To model the disease in vitro, we first engineered the homozygous A>G mutation in H9 ESCs to study the mutation in a patient independent genetic background and compare it to isogenic parental cells. We obtained two independent clones harboring the homozygous A>G change (referred to as knock-in, KI, mutant) and two cell lines harboring an insertion of an additional A after nucleotide position 42 of UGP2 transcript 1 (chr2:64083462_64083463insA) (Supplementary Fig. 3a, b, online resource) (referred to as knockout, KO). This causes a premature stop codon at amino acid position 47 (D15Rfs*33), leading to nonsense-mediated mRNA decay and complete absence of UGP2 protein (Supplementary Fig. 3c, online resource). All derived ESCs had a normal morphology and remained pluripotent as assessed by marker expression (Supplementary Fig. 3d, e, online resource), indicating that the absence of UGP2 in ESCs is tolerated, in agreement with genome-wide LoF CRISPR screens which did not identify UGP2 as an essential gene in ESCs [66, 119]. We differentiated wild type, KI and KO ESCs into NSCs, using dual SMAD inhibition (Supplementary Fig. 4a–c, online resource). Wild-type cells could readily differentiate into NSCs, having a normal morphology and marker expression, whereas differentiation of KI and KO cells was more variable and not all differentiations resulted in viable, proliferating NSCs. KO cells could not be propagated for more than five passages under NSC culture conditions (data not shown), which could indicate that the total absence of UGP2 protein is not tolerated in NSCs. When assessed by Western blotting, total UGP2 protein levels were reduced in KI cells and depleted in KO cells compared to wild type (Supplementary Fig. 4d, e, online resource).
Next, we performed RNA-seq of wild type, KI and KO ESCs and NSCs to assess how depletion of UGP2 upon NSC differentiation would impact the global transcriptome (Fig. 4, Supplementary Fig. 5, Supplementary Table 2, online resource). In agreement with normal proliferation and morphology of KI and KO ESCs, all ESCs shared a similar expression profile of pluripotency-associated genes and only few genes were differentially expressed between the three genotypes (Supplementary Fig. 5c, Supplementary Table 3, online resource). This indicates that the absence of UGP2 in ESCs does not lead to major transcriptome alterations despite the central role of this enzyme in metabolism. Upon differentiation, cells from all genotypes expressed NSC markers (Supplementary Fig. 5f, online resource), but when comparing wild type and KO cells, we observed noticeable changes that were less pronounced in KI NSCs but still followed a similar trend (Fig. 4a, b, Supplementary Fig. 5d, e, online resource). Gene enrichment analysis showed that genes downregulated in KO and KI cells were implicated in processes related to the extracellular matrix, cell–cell interactions and metabolism, while genes upregulated in KO and KI cells were enriched for synaptic processes and genes implicated in epilepsy (Fig. 4c, Supplementary Table 4, online resource). Both KO and KI cells showed an upregulation of neuronal expressed genes, indicating a tendency to differentiate prematurely. To validate RNA-seq findings, we tested several genes by qRT-PCR in wild type, KI and KO cells (Fig. 4d). We also included KO rescue cells, in which we had restored the expression of either the wild type or the mutant UGP2 long isoform, leading each to an approximately fourfold UGP2 overexpression at the NSC state compared to WT (Supplementary Fig. 4f, online resource). Amongst the tested genes was NNAT, which showed a significant upregulation in KI and KO cells, which was rescued by the restoration of UGP2 expression in KO NSCs. NNAT encodes neuronatin that stimulates glycogen synthesis by upregulating glycogen synthase and was previously found to be upregulated in Lafora disease. This lethal teenage-onset neurodegenerative disorder presenting with myoclonic epilepsy is caused by mutations in the ubiquitin ligase malin, leading to accumulation of altered polyglucosans [107]. Malin can ubiquitinate neuronatin leading to its degradation. As reduced UGP2 expression might impact glycogen production, it seems plausible that this results in compensatory NNAT upregulation and in downstream aberrations contributing to the patient phenotypes. Indeed, neuronatin upregulation was shown to cause increased intracellular Ca2+ signaling, ER stress, proteasomal dysfunction and cell death in Lafora disease [92, 93], and was shown to be a stress-responsive protein in the outer segment of retina photoreceptors [91, 95]. Another interesting gene upregulated in KI and KO NSCs and downregulated in rescue cell lines was the autism candidate gene FGFBP3 [87]. This secreted proteoglycan that enhances FGF signaling is broadly expressed in brain [60], and functions as an extracellular chaperone for locally stored FGFs in the ECM, thereby influencing glucose metabolism by regulating rate-limiting enzymes in gluconeogenesis [102]. Other potentially relevant genes displaying the same expression trend were the heparan sulfate proteoglycan GPC2 (a marker of immature neurons [64, 72]), the helix–loop–helix transcription factor ID4 (a marker of postmitotic neurons [29]), and the signaling molecule FGFR3 that has been implicated in epilepsy [73]. Genes downregulated in KO cells and upregulated in rescue cells included urokinase-type plasminogen activator PLAU (deficiency in mouse models increases seizure susceptibility [53]), the glycoprotein GALNT7 (upregulation of which has been found to promote glioma cell invasion [45]) and the brain tumor gene MYBL1 (that has been shown to be regulated by O-linked N-acetylglucosamine [42]. Similar expression changes were observed in NSCs differentiated from induced pluripotent stem cells (iPSCs) that we had generated from family 1 (Supplementary Fig. 6, online resource). Together, RNA-seq showed that whereas the absence of UGP2 is tolerated in ESCs, its complete absence or reduced expression results in global transcriptome changes in NSCs, with many affected genes implicated in DEE-relevant pathways.
RNA-seq of UGP2 mutant H9-derived neural stem cells. a Venn diagram showing the overlap between differentially expressed genes in UGP2 KO or KI NSCs that are upregulated (upper panel, genes with FDR < 0.05 and LogFC > 1) or downregulated (lower panel, genes with FDR < 0.05 and LogFC < -1) compared to wild-type NSCs. b Box plot showing the distribution of gene expression levels [in Log2(RPKM + 1)] from RNA-seq for the groups of genes displayed in a, in wild type, UGP2 KI or KO NSCs. Boxes are IQR; line is median; and whiskers extend to 1.5 × the IQR (*p < 0.05; **p < 0.01, ***p < 0.001, unpaired t test, two tailed). c Enrichment analysis using Enrichr [52] of up- or downregulated genes in NSCs from a for selected gene ontology sets, showing the five most enriched terms per set. Combined score and p value calculated by Enrichr are depicted (*p < 0.05; **p < 0.01; ***p < 0.001). d qRT-PCR validation of differentially expressed genes from RNA-seq in wild type, UGP2 KI, UGP2 KO NSCs and KO NSCs rescued with either WT or MUT (Met12Val) transcript isoform 1, at p5 of NSC differentiation. Bar plot showing the mean fold change for the indicated genes compared to wild type, normalized for the housekeeping gene TBP. Results of two biological and two independent technical replicates are plotted. Colors match the Venn diagram group to which the tested genes belong, from a. Error bars represent SEM; (*p < 0.05; **p < 0.01, ***p < 0.001, unpaired t test, one-tailed)
Absence of short UGP2 isoform leads to metabolic defects in neural stem cells
To investigate how reduced UGP2 expression levels in KO and KI cells would impact NSC metabolism, we investigated the capacity to produce UDP-glucose in the presence of exogenously supplied glucose-1-phosphate and UTP. KO NSCs showed a severely reduced ability to produce UDP-glucose (Fig. 5a). This reduction was rescued by ectopic overexpression of both long wild type and long mutant UGP2. KI cells showed a slightly reduced activity in ESCs (Supplementary Fig. 7a, online resource), but a more strongly reduced activity in NSCs compared to wild type (Fig. 5a), correlating with total UGP2 expression levels (Supplementary Fig. 4d, e, online resource). Surprisingly, contrary to KO NSCs, KO ESC showed some residual capacity to produce UDP-glucose despite the complete absence of UGP2 (Supplementary Fig. 7a, online resource). This could indicate that a yet to be identified enzyme can partially take over the function of UGP2 in ESCs but not NSCs, which might explain the lack of expression changes in this cell type upon UGP2 loss. iPSCs showed similar results (Supplementary Fig. 7b, online resource). We next assessed the capacity to synthesize glycogen under low-oxygen conditions by PAS staining, as it was previously shown that hypoxia triggers increased glycogen synthesis [78]. As expected, wild-type ESCs cultured for 48 h under hypoxia showed an intense cytoplasmic PAS staining in most cells (Supplementary Fig. 7c, d, online resource), while KO ESCs showed a severely reduced staining intensity. This indicates that under hypoxia conditions, the residual capacity of ESC to produce UDP-glucose in the absence of UGP2 is insufficient to produce glycogen. KI ESCs were indistinguishable from wild type (Supplementary Fig. 7d, online resource). At the NSC state, many KO cells kept at low-oxygen conditions for 48 h died (data not shown) and those KO cells that did survive were completely depleted from glycogen granules (Fig. 5b, c). This could be rescued by overexpression of both wild type and mutant long UGP2 isoform. KI NSCs showed a more severe reduction in PAS staining compared to the ESC state (Fig. 5b, c), and we observed similar findings in patient iPSC-derived NSCs (Supplementary Fig. 7e, online resource). Together, this further indicates that upon neural differentiation the isoform expression switch renders patient cells depleted of UGP2, leading to a reduced capacity to synthesize glycogen. This can directly be involved in the DEE phenotype, as, besides affecting energy metabolism, reduction of glycogen in brain has been shown to result in (1) impairment of synaptic plasticity [30]; (2) reduced clearance of extracellular potassium ions leading to neuronal hypersynchronization and seizures [22, 63, 117]; (3) altered glutamate metabolism [88]. To investigate how reduced UDP-glucose levels would impact on glycosylation, we next investigated glycosylation levels by means of LAMP2, a lysosomal protein known to be extensively glycosylated both by N-linked and O-linked glycosylation [114]. We found that KO NSCs show hypoglycosylation of LAMP2 that is rescued by the overexpression of both WT and mutant long isoform (Fig. 5d). In contrast, in ESCs, no glycosylation defects were noticed (Supplementary Fig. 7f, online resource). Finally, we investigated whether the absence of UGP2, affecting protein glycosylation, could induce ER stress and thus unfolded protein response (UPR). Whereas in ESCs, the absence of UGP2 did not result in a detectable effect on UPR markers (Supplementary Fig. 7g, online resource), in NSCs we noticed an increased expression of these genes both in KO and in KI cells (Fig. 5e). This indicates that NSCs having UGP2 levels under a certain threshold are more prone to ER stress and UPR. In agreement with this, we did not observe upregulation of UPR markers in patient-derived fibroblast, which have similar total UGP2 expression levels compared to controls (Supplementary Fig. 7h, online resource). Together this indicates that upon differentiation to NSCs, KI cells become sufficiently depleted of UGP2 to have reduced synthesis of UDP-glucose, leading to defects in glycogen synthesis and protein glycosylation and to the activation of UPR response. Alterations of these crucial processes are likely to be implicated in the pathogenesis leading to increased seizure susceptibility, altered brain microstructure and progressive microcephaly.
Metabolic changes upon UGP2 loss. a UGP2 enzymatic activity in WT, UGP2 KI, KO and KO NSCs rescued with WT or MUT (Met12Val) isoform 1 of UGP2. Bar plot showing the mean of two replicate experiments, error bar is SEM. *p < 0.05; ***p < 0.001, unpaired t test, two tailed. b Representative pictures of PAS staining in WT, KI, KO and rescued NSCs. Nuclei are counterstained with hematoxylin (blue). Inserts show zoom-in of part of the cytoplasm. Note the presence of glycogen granules in WT NSCs, their diminished number in KI NSCs, their absence in KO NSCs and their reappearance upon rescue with WT or MUT (Met12Val) isoform 1 of UGP2. c Quantification of the number of glycogen granules per cell in WT, UGP2 KI, KO and rescued NSCs, after 48 h culture under low-oxygen conditions. Shown is the average number of glycogen granules per cell, n = 80–100 cells per genotype. Error bars represent the SD. ***p < 0.001, unpaired t test, two tailed. d Western blotting detecting LAMP2 (upper panel) and the housekeeping control actin (lower panel) in cellular extracts from H9-derived NSCs that are WT, UGP2 KI, KO and KO cells rescued with WT or MUT (Met12Val) isoform 1 of UGP2. Glycosylated LAMP2 runs at ~ 110 kDa, whereas hypo-glycosylated LAMP2 is detected around 75 kDa. The absence of changes in LAMP2 glycosylation in KI cells is likely explained by a non-complete isoform switch upon in vitro NSC differentiation, resulting in residual UGP2 levels (see Supplementary Fig. 5d, online resource). e qRT-PCR expression analysis for UPR marker genes (spliced XBP1, HSPA5, ATF4 and EDEM) in WT, KI, KO and rescued NSCs. Shown is the mean fold change for the indicated genes compared to wild type, normalized for the housekeeping gene TBP. Results of two biological and two independent technical replicates are plotted, from two experiments. Error bars represent SEM; *p < 0.05; **p < 0.01, ***p < 0.001, unpaired t test, two tailed
ugp2a and ugp2b double mutant zebrafish recapitulate metabolic changes during brain development, have an abnormal behavioral phenotype, visual disturbance, and increased seizure susceptibility
Finally, to model the consequences of the lack of UGP2 in vivo, we generated zebrafish mutants for both ugp2a and ugp2b, the zebrafish homologs of UGP2, using CRISPR–Cas9 injections in fertilized oocytes in a background of a radial glia/neural stem cell reporter [51]. Double homozygous mutant lines having frameshift deletions for both genes confirmed by Sanger sequencing could be generated but the only viable combination, obtained with ugp2a loss, created a novel ATG in exon 2 of ugp2b, leading to a hypomorphic allele (Fig. 6a). Homozygous ugp2a/b mutant zebrafish had a normal gross morphology of brain and radial glial cells (Fig. 6b), showed a largely diminished activity to produce UDP-glucose in the presence of exogenously supplied glucose-1-phosphate and UTP (Fig. 6c), and showed a reduction in c-fos expression levels, indicating reduced global neuronal activity (Fig. 6d). To monitor possible spontaneous seizures, we performed video tracking experiments of developing larvae under light–dark cycling conditions at 5 days post-fertilization (dpf). Control larvae show increased locomotor activity under light conditions, and although ugp2 double mutant larvae still responded to increasing light conditions, they showed a strongly reduced activity (Fig. 6e, f). This could indicate that their capability to sense visual cues is diminished, or that their tectal processing of visual input is delayed, resulting in reduced movements. Strikingly, upon careful inspection, we noticed that ugp2 double mutant larvae did not show spontaneous eye movements, in contrast to age-matched control larvae (Fig. 6g, Supplemental Movies 2 and 3, online resource). Whereas we did not observe an obvious spontaneous epilepsy phenotype in these double mutant larvae, upon stimulation with 4-aminopyridine (4-AP), a potent convulsant, double mutant larvae showed an increased frequency and duration of movements at high velocity compared to controls, which might indicate an increased seizure susceptibility (Fig. 6h, i). Taken together, severely reduced Ugp2a/Ugp2b levels result in a behavior defect with reduced eye movements, indicating that also in zebrafish Ugp2 plays an important role in brain function.
Zebrafish disease modeling. a Schematic drawing of the ugp2a and ugp2b loci in zebrafish and the generated mutations. b Confocal images (maximum projection of confocal Z-stacks) of the brain of wild type (left) and ugp2aΔ/Δ; ugp2bΔ/Δ mutant zebrafish larvae (right), both in an slc1a2b-citrine reporter background, at 4 days post-fertilization (dpf). The lower panels are higher magnifications of the boxed regions indicated in the upper panels. Scale bar in upper panel is 100 µm, in lower panel 20 µm. In upper panel, Z = 45 with step size 4 µm; in lower panel, Z = 30 with step size 2 µm. c Enzymatic activity in ugp2 double mutant zebrafish larvae at 4 and 5 dpf, compared to wild-type age-matched controls, showing reduced Ugp2 enzyme activity in double mutant zebrafish. d qRT-PCR for the neuronal activity marker c-fos in wild type and ugp2 double mutant larvae at 3 dpf. For each group, 2 batches of 12 larvae were pooled. Shown is the mean fold change for the indicated genes compared to wild type, normalized for the housekeeping gene gapdh. Error bars represent SEM; ***p < 0.001, unpaired t test, two tailed. e Representative graph of a locomotion assay showing the total distance moved by larvae during the dusk–dawn routine (total time: 3 h 12 min), n = 24 larvae per genotype. Gray shading shows the standard error of the mean. f Quantification of the total distance moved throughout the experiment from e excluding the dark period. g Quantification of the number of observed spontaneous eye movements during a 2-min observation in wild type and ugp2 double mutant larvae at 4 dpf. Each dot represents one larva; shown is the average and SD; ***p < 0.001, t test, two tailed. h Quantification of the frequency of movements at a speed of > 15 mm/s, for wild-type control and ugp2 double mutant zebrafish larvae at 4 dpf, treated with mock control or with 0.04 nM or 0.4 nM 4-AP during a 35-min observation. Each dot represents a single larva; results of two experiments are shown, within total 24 larvae per condition. i Quantification of the movement duration at a speed of > 15 mm/s, for wild-type control and ugp2 double mutant zebrafish larvae at 4 dpf, treated with mock control or with 0.04 nM or 0.4 nM 4-AP during a 35-min observation. Each dot represents a single larva; results of two experiments are shown, with in total 24 larvae per condition. *p < 0.05, two-way ANOVA with Bonferoni post test
UGP2 is an essential gene in humans and ATG mutations of tissue-specific isoforms of essential genes potentially cause more rare genetic diseases
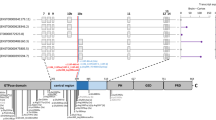
Several lines of evidence argue that UGP2 is essential in humans. First, no homozygous LoF variants or homozygous exon-covering deletions for UGP2 are present in gnomAD or GeneDx controls, and homozygous variants in this gene are limited to non-coding changes, synonymous variants and five missense variants, together occurring only seven times homozygous (Supplementary Table 5, online resource). Also, no homozygous or compound heterozygous UGP2 LoF variants were found in published studies on dispensable genes in human knockouts [70, 86, 99], or in the Centogene (CentoMD®) or GeneDx patient cohorts, encompassing together many thousands of individuals, further indicating that this gene is intolerant to loss-of-function in a bi-allelic state. In addition, no homozygous deletions of the region encompassing UGP2 are present in DECIPHER [35] or ClinVar [54]. Second, UGP2 has been identified as an essential gene using gene-trap integrations [17] and in CRISPR–Cas9 LoF screens in several human cell types [5, 15, 43, 113, 115]. Finally, studies in yeast [25, 26], fungus [58] and plants [21, 75, 116] consider the orthologs of UGP2 as essential, and the absence of Ugp2 in mice is predicted to be lethal [104]. In flies, homozygous UGP knockouts are lethal while only hypomorphic compound heterozygous alleles are viable but have a severe movement defect with altered neuromuscular synaptogenesis due to glycosylation defects [48]. To further investigate the essentiality of UGP2, we performed differentiation experiments of our WT, KO and rescue ESCs. Differentiation of KO ESCs into hematopoietic stem cells (HSCs) resulted in severe downregulation of GATA2 compared to wild-type cells, and this was restored in rescue cell lines (Fig. 7a). GATA2 is a key transcription factor in the developing blood system, and knockout of Gata2 is embryonic lethal in mice due to defects in HSC generation and maintenance [28, 106]. Differentiation of ESCs into cardiomyocytes similarly affected key marker gene expression in KO cells, and these changes were restored upon UGP2 rescue (Fig. 7b, c). Whereas WT ESCs could generate beating cardiomyocytes after 10 days, these were not seen in KO ESCs. Taken together this argues that the complete absence of UGP2 in humans is probably incompatible with life, a hypothesis that cannot be tested directly. However, if true, this could well explain the occurrence of the unique recurrent mutation in all cases presented herein. Given the structure of the UGP2 locus (Fig. 2a), every LoF variant would affect either the long isoform, when located in the first 33 nucleotides of the cDNA sequence, or both the short and long isoforms when downstream to the ATG of the short isoform. Therefore, the short isoform start codon is the only mutational target that can disrupt specifically the short isoform. In this case, the Met12Val change introduced into the long isoform does not seem to disrupt UGP2 function to such an extent that this is intolerable and, therefore, allows development to proceed for most tissues. However, the lack of the short UGP2 isoform caused by the start codon mutation results in a depletion of functional UGP2 in tissues where normally the short isoform is predominantly expressed. In brain, this reduction diminishes total UGP2 levels below a threshold for normal development, causing a severe epileptic encephalopathy syndrome. Given the complexity of the human genome with 42,976 transcripts with RefSeq peptide IDs, perhaps also other genetic disorders might be caused by such tissue-restricted depletion of essential proteins. Using a computational homology search of human proteins encoded by different isoforms, we have identified 1766 genes that share a similar structure to the UGP2 locus (e.g., a shorter protein isoform that is largely identical to the longer protein isoform, translated from an ATG that is contained within the coding sequence of the long isoform) (Fig. 7d). When filtering these genes for (1) those previously shown to be essential [10], (2) not associated with disease (e.g., no OMIM phenotype) and (3) those proteins where the shorter isoform is no more than 50 amino acids truncated at the N-terminal compared to the longer isoform, we identified 247 genes (Supplementary Table 6, online resource). When comparing the ratios of isoform-specific reads obtained from different fetal RNA-seq data [46, 83, 94, 118], we noticed that many of these genes show differential isoform expression amongst multiple tissues, with many genes showing either expression of the long or the short isoform in a particular tissue (Fig. 7e). Homozygous LoF variants or start codon altering mutations in these genes are rare in gnomAD (Supplementary Table 7, online resource), and it is tempting to speculate that mutations in start codons of these genes could be associated with human genetic diseases, as is the case for UGP2. Using mining of data from undiagnosed patients from our own exome database, the Queen Square Genomic Center database and those from Centogene and GeneDx, we found evidence for several genes out of the 247 having rare, bi-allelic variants affecting the start codon of one of the isoforms that could be implicated in novel disorders (unpublished observations) and give one such example in the Supplementary Note, online resource. Together, these findings highlight the relevance of mutations resulting in tissue-specific protein loss of essential genes for genetic disorders.
Essentiality of UGP2 and other disease candidate genes with a similar mutation mechanism. a qRT-PCR analysis of the hematopoietic stem cell markers GATA2, LMO2 and RUNX1, after 12 days of differentiation of wild type, UGP2 KO and UGP2 KO rescue ESCs. Shown is the mean fold change for the indicated genes compared to wild type, normalized for the housekeeping gene TBP. Results of two biological and two technical replicates are plotted. Error bars represent SEM; *p < 0.05; **p < 0.01, ***p < 0.001, unpaired t test, two tailed. b As a, but now for cardiomyocyte differentiation at day 15, assessing expression of the cardiomyocyte markers TNNT2, MYL2 and MYL7. c Bright-field image of cardiomyocyte cultures of wild type, UGP2 KO and rescue cells. Note the elongated organized monolayer structure cardiomyocytes capable of beating in wild type and rescue cells that are absent in KO cultures. Scale bar is 400 µm. d Scheme showing the homology search to identify genes with a similar structure as UGP2, where ATG-altering mutations could affect a tissue-specific isoform causing genetic disease. e Heat map showing the ratio of short isoform expression over total isoform expression from published RNA-seq data amongst 20 tissues for 83 out 247 essential genes that are not yet implicated in disease and in which the short and longer protein isoforms differ by less than 50 amino acids at the N-terminal
Discussion
Here we describe a recurrent variant in 22 individuals from 15 families, affecting the start codon of the shorter isoform of the essential gene UGP2 as a novel cause of a severe DEE. Using in vitro and in vivo disease modeling, we provide evidence that the reduction of UGP2 expression in brain cells leads to global transcriptome changes, a reduced ability to produce glycogen, alterations in glycosylation and increased sensitivity to ER stress, which together can explain the phenotype observed in the patients. Most likely our findings in vitro underestimate the downstream effects in patient cells, as in fetal brain, the longer isoform expression is almost completely silenced and virtually all UGP2 come from the shorter isoform, which in patient cells cannot be translated. During our in vitro NSC differentiation, this isoform switch is less complete, leaving cells with the patient mutation with some residual UGP2. Strikingly, the clinical phenotype seems to be very similar in all cases, including intractable seizures, absence of developmental milestones, progressive microcephaly and a disturbance of vision, with retinal pigment changes observed in all patients who had undergone ophthalmological examination. Also, all patients seem to share similar, although mild, dysmorphisms, possibly making this condition a recognizable syndrome.
The involvement of UGP2 in genetic disease is surprising. Given its central role in nucleotide-sugar metabolism it is expected that loss of this essential protein would be incompatible with life and, therefore, loss-of-function should not be found in association with postnatal disease. Our data argue that indeed a total absence of UGP2 in all cells is lethal, but that tissue-specific loss, as caused here by the start codon alteration of an isoform important for brain, can be compatible with postnatal development but still results in a severe phenotype. Given that any other LoF variant across this gene would most likely affect both protein isoforms, this could also explain why only a single mutation is found in all individuals. The fact that the Met12Val long isoform was able to rescue the full KO phenotype indicates that the missense change introduced to the long protein isoform does not affect UGP2 function. As other variants at this start codon, even heterozygous, are not found, possibly missense variants encoding for leucine, lysine, threonine, arginine or isoleucine (e.g., amino acids that would be encoded by alternative changes affecting the ATG codon) at this amino acid location in the long isoform could not produce a functional protein and are, therefore, not tolerated. Although start codon mutations have previously been implicated in disease [16, 19], there are no reports, to our knowledge, on disorders describing start codon alterations of other essential genes, leading to alterations of tissue-specific isoforms. Using a genome-wide homology search, we have identified a large list of other essential genes with a similar locus structure and variable isoform expression amongst tissues, where similar ATG-altering variants could affect tissue-relevant expression. An intriguing question is why evolution has resulted in a large number of genes encoding almost identical protein isoforms. It will be interesting to further explore the mutational landscape of these genes in cohorts of currently unexplained patients.
References
(2010) Baluchistan i. Geography, history and ethnography. Encyclopædia Iranica City, pp fasc. 6, pp 598–632
Adeva-Andany MM, Gonzalez-Lucan M, Donapetry-Garcia C, Fernandez-Fernandez C, Ameneiros-Rodriguez E (2016) Glycogen metabolism in humans. BBA Clin 5:85–100
Anders S, Pyl PT, Huber W (2015) HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169
Arranz AM, Perkins KL, Irie F, Lewis DP, Hrabe J, Xiao F et al (2014) Hyaluronan deficiency due to Has3 knock-out causes altered neuronal activity and seizures via reduction in brain extracellular space. J Neurosci 34:6164–6176
Bakke J, Wright WC, Zamora AE, Oladimeji P, Crawford JC, Brewer CT et al (2019) Genome-wide CRISPR screen reveals PSMA6 to be an essential gene in pancreatic cancer cells. BMC Cancer 19:253
Barakat TS, Ghazvini M, de Hoon B, Li T, Eussen B, Douben H et al (2015) Stable X chromosome reactivation in female human induced pluripotent stem cells. Stem Cell Rep 4:199–208
Barakat TS, Gribnau J (2015) Generation of knockout alleles by RFLP based BAC targeting of polymorphic embryonic stem cells. Methods Mol Biol 1227:143–180
Barakat TS, Gribnau J (2010) X chromosome inactivation and embryonic stem cells. Adv Exp Med Biol 695:132–154
Barakat TS, Halbritter F, Zhang M, Rendeiro AF, Perenthaler E, Bock C et al (2018) Functional dissection of the enhancer repertoire in human embryonic stem cells. Cell Stem Cell 23(276–288):e278
Bartha I, di Iulio J, Venter JC, Telenti A (2018) Human gene essentiality. Nat Rev Genet 19:51–62
Bayer SA, Altman J (2004) Atlas of human central nervous system development: the human brain during the third trimester, vol 2. CRC Press, New York
Bayer SA, Altman J (2005) Atlas of human central nervous system development: the human brain during the second trimester, vol 3. CRC Press, New York
Bayer SA, Altman J (2006) Atlas of human central nervous system development: the human brain during the late first trimester, vol 4. CRC Press, New York
Bayer SA, Altman J (2008) Atlas of human central nervous system development: the human brain during the early first trimester, vol 5. CRC Press, New York
Bertomeu T, Coulombe-Huntington J, Chatr-Aryamontri A, Bourdages KG, Coyaud E, Raught B et al (2018) A high-resolution genome-wide CRISPR/Cas9 viability screen reveals structural features and contextual diversity of the human cell-essential proteome. Mol Cell Biol 38:10
Binder J, Hofmann S, Kreisel S, Wohrle JC, Bazner H, Krauss JK et al (2003) Clinical and molecular findings in a patient with a novel mutation in the deafness-dystonia peptide (DDP1) gene. Brain 126:1814–1820
Blomen VA, Majek P, Jae LT, Bigenzahn JW, Nieuwenhuis J, Staring J et al (2015) Gene essentiality and synthetic lethality in haploid human cells. Science 350:1092–1096
Brinkman EK, Chen T, Amendola M, van Steensel B (2014) Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 42:e168
Caridi G, Dagnino M, Lugani F, Shalev SA, Campagnoli M, Galliano M et al (2013) A novel mutation in the albumin gene (c.1A %3e C) resulting in analbuminemia. Eur J Clin Invest 43:72–78
Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L (2009) Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 27:275–280
Chen R, Zhao X, Shao Z, Wei Z, Wang Y, Zhu L et al (2007) Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility. Plant Cell 19:847–861
Choi HB, Gordon GR, Zhou N, Tai C, Rungta RL, Martinez J et al (2012) Metabolic communication between astrocytes and neurons via bicarbonate-responsive soluble adenylyl cyclase. Neuron 75:1094–1104
Consortium GT (2013) The genotype-tissue expression (GTEx) project. Nat Genet 45:580–585
Cope EC, Gould E (2019) Adult Neurogenesis, glia, and the extracellular matrix. Cell Stem Cell 24(5):690–705
Daran JM, Bell W, Francois J (1997) Physiological and morphological effects of genetic alterations leading to a reduced synthesis of UDP-glucose in Saccharomyces cerevisiae. FEMS Microbiol Lett 153:89–96
Daran JM, Dallies N, Thines-Sempoux D, Paquet V, Francois J (1995) Genetic and biochemical characterization of the UGP1 gene encoding the UDP-glucose pyrophosphorylase from Saccharomyces cerevisiae. Eur J Biochem 233:520–530
de Jonge HJ, Woolthuis CM, Vos AZ, Mulder A, van den Berg E, Kluin PM et al (2011) Gene expression profiling in the leukemic stem cell-enriched CD34 + fraction identifies target genes that predict prognosis in normal karyotype AML. Leukemia 25:1825–1833
de Pater E, Kaimakis P, Vink CS, Yokomizo T, Yamada-Inagawa T, van der Linden R et al (2013) Gata2 is required for HSC generation and survival. J Exp Med 210:2843–2850
Diotel N, Beil T, Strahle U, Rastegar S (2015) Differential expression of id genes and their potential regulator znf238 in zebrafish adult neural progenitor cells and neurons suggests distinct functions in adult neurogenesis. Gene Expr Patterns 19:1–13
Duran J, Saez I, Gruart A, Guinovart JJ, Delgado-Garcia JM (2013) Impairment in long-term memory formation and learning-dependent synaptic plasticity in mice lacking glycogen synthase in the brain. J Cereb Blood Flow Metab 33:550–556
Epi25 Collaborative, Electronic address sbuea, Epi C (2019) Ultra-rare genetic variation in the epilepsies: a whole-exome sequencing study of 17,606 individuals. Am J Hum Genet 1:4
Epi KC, Phenome E, Genome P, Allen AS, Berkovic SF, Cossette P, Delanty N et al (2013) De novo mutations in epileptic encephalopathies. Nature 501:217–221
Exome Variant Server NHLBI GO Exome Sequencing Project (ESP) Seattle WA (accessed Juli 2019).
Fattahi Z, Beheshtian M, Mohseni M, Poustchi H, Sellars E, Nezhadi SH et al (2019) Iranome: a catalog of genomic variations in the Iranian population. Hum Mutat 1:4
Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D et al (2009) DECIPHER: database of chromosomal imbalance and phenotype in humans using Ensembl resources. Am J Hum Genet 84:524–533
Flores-Diaz M, Alape-Giron A, Persson B, Pollesello P, Moos M, von Eichel-Streiber C et al (1997) Cellular UDP-glucose deficiency caused by a single point mutation in the UDP-glucose pyrophosphorylase gene. J Biol Chem 272:23784–23791
Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT (2011) LOVD vol 2.0: the next generation in gene variant databases. Hum Mutat 32:557–563
Fuhring J, Damerow S, Fedorov R, Schneider J, Munster-Kuhnel AK, Gerardy-Schahn R (2013) Octamerization is essential for enzymatic function of human UDP-glucose pyrophosphorylase. Glycobiology 23:426–437
Fuhring JI, Cramer JT, Schneider J, Baruch P, Gerardy-Schahn R, Fedorov R (2015) A quaternary mechanism enables the complex biological functions of octameric human UDP-glucose pyrophosphorylase, a key enzyme in cell metabolism. Sci Rep 5:9618
Gonzalez M, Falk MJ, Gai X, Postrel R, Schule R, Zuchner S (2015) Innovative genomic collaboration using the GENESIS (GEM.app) platform. Hum Mutat 36:950–956
Gostynska KB, Nijenhuis M, Lemmink H, Pas HH, Pasmooij AM, Lang KK et al (2015) Mutation in exon 1a of PLEC, leading to disruption of plectin isoform 1a, causes autosomal-recessive skin-only epidermolysis bullosa simplex. Hum Mol Genet 24:3155–3162
Guo H, Zhang B, Nairn AV, Nagy T, Moremen KW, Buckhaults P et al (2017) O-Linked N-acetylglucosamine (O-GlcNAc) expression levels epigenetically regulate colon cancer tumorigenesis by affecting the cancer stem cell compartment via modulating expression of transcriptional factor MYBL1. J Biol Chem 292:4123–4137
Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G et al (2015) High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163:1515–1526
Higuita JC, Alape-Giron A, Thelestam M, Katz A (2003) A point mutation in the UDP-glucose pyrophosphorylase gene results in decreases of UDP-glucose and inactivation of glycogen synthase. Biochem J 370:995–1001
Hua S, Li H, Liu Y, Zhang J, Cheng Y, Dai C (2018) High expression of GALNT7 promotes invasion and proliferation of glioma cells. Oncol Lett 16:6307–6314
Hwang T, Park CK, Leung AK, Gao Y, Hyde TM, Kleinman JE et al (2016) Dynamic regulation of RNA editing in human brain development and disease. Nat Neurosci 19:1093–1099
Jenkins ZA, Macharg A, Chang CY, van Kogelenberg M, Morgan T, Frentz S et al (2018) Differential regulation of two FLNA transcripts explains some of the phenotypic heterogeneity in the loss-of-function filaminopathies. Hum Mutat 39:103–113
Jumbo-Lucioni PP, Parkinson WM, Kopke DL, Broadie K (2016) Coordinated movement, neuromuscular synaptogenesis and trans-synaptic signaling defects in Drosophila galactosemia models. Hum Mol Genet 25:3699–3714
Kalser J, Cross JH (2018) The epileptic encephalopathy jungle—from Dr West to the concepts of aetiology-related and developmental encephalopathies. Curr Opin Neurol 31:216–222
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360
Kuil LE, Lopez Marti A, Carreras Mascaro A, van den Bosch JC, van den Berg P, van der Linde HC et al (2019) Hexb enzyme deficiency leads to lysosomal abnormalities in radial glia and microglia in zebrafish brain development. Glia 67:1705–1718
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z et al (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44:W90–97
Kyyriainen J, Bolkvadze T, Koivisto H, Lipponen A, Perez LO, Ekolle Ndode-Ekane X et al (2019) Deficiency of urokinase-type plasminogen activator and its receptor affects social behavior and increases seizure susceptibility. Epilepsy Res 151:67–74
Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM et al (2014) ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42:D980–985
Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R et al (2013) Software for computing and annotating genomic ranges. PLoS Comput Biol 9:e1003118
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T et al (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536:285–291
Li J, Woods SL, Healey S, Beesley J, Chen X, Lee JS et al (2016) Point mutations in exon 1B of APC reveal gastric adenocarcinoma and proximal polyposis of the stomach as a familial adenomatous polyposis variant. Am J Hum Genet 98:830–842
Li M, Chen T, Gao T, Miao Z, Jiang A, Shi L et al (2015) UDP-glucose pyrophosphorylase influences polysaccharide synthesis, cell wall components, and hyphal branching in Ganoderma lucidum via regulation of the balance between glucose-1-phosphate and UDP-glucose. Fungal Genet Biol 82:251–263
Li S, Hu Z, Zhao Y, Huang S, He X (2019) Transcriptome-wide analysis reveals the landscape of aberrant alternative splicing events in liver cancer. Hepatology 69:359–375
Li Y, Sun S, Ding Z, Yang C, Zhang G, Jiang Q et al (2018) Temporal and spatial expression of fgfbp genes in zebrafish. Gene 659:128–136
Li Y, Zhuang H, Zhang X, Li Y, Liu Y, Yi X et al (2018) Multiomics integration reveals the landscape of prometastasis metabolism in hepatocellular carcinoma. Mol Cell Proteom 17:607–618
Lindhout M, Rubio-Gozalbo ME, Bakker JA, Bierau J (2010) Direct non-radioactive assay of galactose-1-phosphate:uridyltransferase activity using high performance liquid chromatography. Clin Chim Acta 411:980–983
Lopez-Ramos JC, Duran J, Gruart A, Guinovart JJ, Delgado-Garcia JM (2015) Role of brain glycogen in the response to hypoxia and in susceptibility to epilepsy. Front Cell Neurosci 9:431
Lugert S, Kremer T, Jagasia R, Herrmann A, Aigner S, Giachino C et al (2017) Glypican-2 levels in cerebrospinal fluid predict the status of adult hippocampal neurogenesis. Sci Rep 7:46543
Magee C, Nurminskaya M, Linsenmayer TF (2001) UDP-glucose pyrophosphorylase: up-regulation in hypertrophic cartilage and role in hyaluronan synthesis. Biochem J 360:667–674
Mair B, Tomic J, Masud SN, Tonge P, Weiss A, Usaj M et al (2019) Essential gene profiles for human pluripotent stem cells identify uncharacterized genes and substrate dependencies. Cell Rep 27(2):599–615.e12
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
McTague A, Howell KB, Cross JH, Kurian MA, Scheffer IE (2016) The genetic landscape of the epileptic encephalopathies of infancy and childhood. Lancet Neurol 15:304–316
Mencacci NE, Kamsteeg EJ, Nakashima K, R'Bibo L, Lynch DS, Balint B et al (2016) De novo mutations in PDE10A cause childhood-onset chorea with bilateral striatal lesions. Am J Hum Genet 98:763–771
Narasimhan VM, Hunt KA, Mason D, Baker CL, Karczewski KJ, Barnes MR et al (2016) Health and population effects of rare gene knockouts in adult humans with related parents. Science 352:474–477
Nashabat M, Al Qahtani XS, Almakdob S, Altwaijri W, Ba-Armah DM, Hundallah K et al (2019) The landscape of early infantile epileptic encephalopathy in a consanguineous population. Seizure 69:154–172
Oikari LE, Okolicsanyi RK, Qin A, Yu C, Griffiths LR, Haupt LM (2016) Cell surface heparan sulfate proteoglycans as novel markers of human neural stem cell fate determination. Stem Cell Res 16:92–104
Okazaki T, Saito Y, Ueda R, Awashima T, Nishimura Y, Yuasa I et al (2017) Epileptic phenotype of FGFR3-related bilateral medial temporal lobe dysgenesis. Brain Dev 39:67–71
Papuc SM, Abela L, Steindl K, Begemann A, Simmons TL, Schmitt B et al (2019) The role of recessive inheritance in early-onset epileptic encephalopathies: a combined whole-exome sequencing and copy number study. Eur J Hum Genet 27:408–421
Park JI, Ishimizu T, Suwabe K, Sudo K, Masuko H, Hakozaki H et al (2010) UDP-glucose pyrophosphorylase is rate limiting in vegetative and reproductive phases in Arabidopsis thaliana. Plant Cell Physiol 51:981–996
Perenthaler E, Yousefi S, Niggl E, Barakat TS (2019) Beyond the exome: the non-coding genome and enhancers in neurodevelopmental disorders and malformations of cortical development. Front Cell Neurosci 13:352
Perkins KL, Arranz AM, Yamaguchi Y, Hrabetova S (2017) Brain extracellular space, hyaluronan, and the prevention of epileptic seizures. Rev Neurosci 28:869–892
Pescador N, Villar D, Cifuentes D, Garcia-Rocha M, Ortiz-Barahona A, Vazquez S et al (2010) Hypoxia promotes glycogen accumulation through hypoxia inducible factor (HIF)-mediated induction of glycogen synthase 1. PLoS ONE 5:e9644
Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H et al (2019) g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 47:W191–W198
Renaud JB, Boix C, Charpentier M, De Cian A, Cochennec J, Duvernois-Berthet E et al (2016) Improved genome editing efficiency and flexibility using modified oligonucleotides with TALEN and CRISPR-Cas9 nucleases. Cell Rep 14:2263–2272
Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M (2019) CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res 47:D886–D894
Retterer K, Juusola J, Cho MT, Vitazka P, Millan F, Gibellini F et al (2016) Clinical application of whole-exome sequencing across clinical indications. Genet Med 18:696–704
Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A et al (2015) Integrative analysis of 111 reference human epigenomes. Nature 518:317–330
Robbins SM, Thimm MA, Valle D, Jelin AC (2019) Genetic diagnosis in first or second trimester pregnancy loss using exome sequencing: a systematic review of human essential genes. J Assist Reprod Genet 36:1539–1548
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Saleheen D, Natarajan P, Armean IM, Zhao W, Rasheed A, Khetarpal SA et al (2017) Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature 544:235–239
Salyakina D, Cukier HN, Lee JM, Sacharow S, Nations LD, Ma D et al (2011) Copy number variants in extended autism spectrum disorder families reveal candidates potentially involved in autism risk. PLoS ONE 6:e26049
Schousboe A, Sickmann HM, Walls AB, Bak LK, Waagepetersen HS (2010) Functional importance of the astrocytic glycogen-shunt and glycolysis for maintenance of an intact intra/extracellular glutamate gradient. Neurotox Res 18:94–99
Schwarz JM, Cooper DN, Schuelke M, Seelow D (2014) MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 11:361–362
Scott EM, Halees A, Itan Y, Spencer EG, He Y, Azab MA et al (2016) Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat Genet 48:1071–1076
Sel S, Patzel E, Poggi L, Kaiser D, Kalinski T, Schicht M et al (2017) Temporal and spatial expression pattern of Nnat during mouse eye development. Gene Express Patterns 23–24:7–12
Sharma J, Mukherjee D, Rao SN, Iyengar S, Shankar SK, Satishchandra P et al (2013) Neuronatin-mediated aberrant calcium signaling and endoplasmic reticulum stress underlie neuropathology in Lafora disease. J Biol Chem 288:9482–9490
Sharma J, Rao SN, Shankar SK, Satishchandra P, Jana NR (2011) Lafora disease ubiquitin ligase malin promotes proteasomal degradation of neuronatin and regulates glycogen synthesis. Neurobiol Dis 44:133–141
Shih HP, Seymour PA, Patel NA, Xie R, Wang A, Liu PP et al (2015) A gene regulatory network cooperatively controlled by Pdx1 and Sox9 governs lineage allocation of foregut progenitor cells. Cell Rep 13:326–336
Shinde V, Pitale PM, Howse W, Gorbatyuk O, Gorbatyuk M (2016) Neuronatin is a stress-responsive protein of rod photoreceptors. Neuroscience 328:1–8
Snoeijen-Schouwenaars FM, van Ool JS, Verhoeven JS, van Mierlo P, Braakman HMH, Smeets EE et al (2019) Diagnostic exome sequencing in 100 consecutive patients with both epilepsy and intellectual disability. Epilepsia 60:155–164
Sobreira N, Schiettecatte F, Valle D, Hamosh A (2015) GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat 36:928–930
Soleman S, Filippov MA, Dityatev A, Fawcett JW (2013) Targeting the neural extracellular matrix in neurological disorders. Neuroscience 253:194–213
Sulem P, Helgason H, Oddson A, Stefansson H, Gudjonsson SA, Zink F et al (2015) Identification of a large set of rare complete human knockouts. Nat Genet 47:448–452
Ta-Shma A, Khan TN, Vivante A, Willer JR, Matak P, Jalas C et al (2017) Mutations in TMEM260 cause a pediatric neurodevelopmental, cardiac, and renal syndrome. Am J Hum Genet 100:666–675
Tan GS, Lim KH, Tan HT, Khoo ML, Tan SH, Toh HC et al (2014) Novel proteomic biomarker panel for prediction of aggressive metastatic hepatocellular carcinoma relapse in surgically resectable patients. J Proteome Res 13:4833–4846
Tassi E, Garman KA, Schmidt MO, Ma X, Kabbara KW, Uren A et al (2018) Fibroblast growth factor binding protein 3 (FGFBP3) impacts carbohydrate and lipid metabolism. Sci Rep 8:15973
Thorsen K, Schepeler T, Oster B, Rasmussen MH, Vang S, Wang K et al (2011) Tumor-specific usage of alternative transcription start sites in colorectal cancer identified by genome-wide exon array analysis. BMC Genom 12:505
Tian D, Wenlock S, Kabir M, Tzotzos G, Doig AJ, Hentges KE (2018) Identifying mouse developmental essential genes using machine learning. Dis Model Mech 11:4
Trujillano D, Bertoli-Avella AM, Kumar Kandaswamy K, Weiss ME, Koster J, Marais A et al (2017) Clinical exome sequencing: results from 2819 samples reflecting 1000 families. Eur J Hum Genet 25:176–182
Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M et al (1994) An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 371:221–226
Turnbull J, Tiberia E, Striano P, Genton P, Carpenter S, Ackerley CA et al (2016) Lafora disease. Epilept Disord 18:38–62
Turnquist RL, Gillett TA, Hansen RG (1974) Uridine diphosphate glucose pyrophosphorylase. Crystallization and properties of the enzyme from rabbit liver and species comparisons. J Biol Chem 249:7695–7700
Turton KB, Esnault S, Delain LP, Mosher DF (2016) Merging absolute and relative quantitative PCR data to quantify STAT3 splice variant transcripts. J Vis Exp 1:4
Vigetti D, Viola M, Karousou E, De Luca G, Passi A (2014) Metabolic control of hyaluronan synthases. Matrix Biol 35:8–13
Wang L, Xiong L, Wu Z, Miao X, Liu Z, Li D et al (2018) Expression of UGP2 and CFL1 expression levels in benign and malignant pancreatic lesions and their clinicopathological significance. World J Surg Oncol 16:11
Wang Q, Yang ZL, Zou Q, Yuan Y, Li J, Liang L et al (2016) SHP2 and UGP2 are biomarkers for progression and poor prognosis of gallbladder cancer. Cancer Invest 34:255–264
Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ et al (2015) Identification and characterization of essential genes in the human genome. Science 350:1096–1101
Wang X, Ju L, Fan J, Zhu Y, Liu X, Zhu K et al (2014) Histone H3K4 methyltransferase Mll1 regulates protein glycosylation and tunicamycin-induced apoptosis through transcriptional regulation. Biochim Biophys Acta 1843:2592–2602
Wang X, Wang S, Troisi EC, Howard TP, Haswell JR, Wolf BK et al (2019) BRD9 defines a SWI/SNF sub-complex and constitutes a specific vulnerability in malignant rhabdoid tumors. Nat Commun 10:1881
Woo MO, Ham TH, Ji HS, Choi MS, Jiang W, Chu SH et al (2008) Inactivation of the UGPase1 gene causes genic male sterility and endosperm chalkiness in rice (Oryza sativa L.). Plant J 54:190–204
Xu J, Song D, Xue Z, Gu L, Hertz L, Peng L (2013) Requirement of glycogenolysis for uptake of increased extracellular K + in astrocytes: potential implications for K + homeostasis and glycogen usage in brain. Neurochem Res 38:472–485
Yan L, Guo H, Hu B, Li R, Yong J, Zhao Y et al (2016) Epigenomic landscape of human fetal brain, heart, and liver. J Biol Chem 291:4386–4398
Yilmaz A, Peretz M, Aharony A, Sagi I, Benvenisty N (2018) Defining essential genes for human pluripotent stem cells by CRISPR-Cas9 screening in haploid cells. Nat Cell Biol 20:610–619
Ying D, Sham PC, Smith DK, Zhang L, Lau YL, Yang W (2015) HaploShare: identification of extended haplotypes shared by cases and evaluation against controls. Genome Biol 16:92
Yu Q, Zheng X (2012) The crystal structure of human UDP-glucose pyrophosphorylase reveals a latch effect that influences enzymatic activity. Biochem J 442:283–291
Zeng C, Xing W, Liu Y (2019) Identification of UGP2 as a progression marker that promotes cell growth and motility in human glioma. J Cell Biochem 120:12489–12499
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O et al (2019) Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10:1523
Acknowledgements
We are indebted to the parents of the patients for their kind cooperation. We thank Virginie Verhoeven and Gerben Schaaf for critically reading our manuscript and Grazia Mancini for helpful discussions. We thank Gerben Schaaf for providing the LAMP2 antibody, and Eskeatnaf Mulugeta for bioinformatics advice. We would like to thank Reviewer 1 for proposing the name “Barakat-Perenthaler-syndrome of developmental epileptic encephalopathy” for this new disorder. DP was supported by an Erasmus + Traineeship Programme. MAS was supported by the King Saud University (RSP-2019/38). AGES was supported by the Yale Center for Mendelian Genomics (NIH Grant M#UM1HG006504-05). HH is supported by the Rosetree Trust, Ataxia UK, MSA Trust, Brain Research UK, Muscular Dystrophy UK, Muscular Dystrophy Association, Higher Education Commission of Pakistan, The MRC (MR/S01165X/1, MR/S005021/1, G0601943), Wellcome Trust (WT093205MA, WT104033AIA, Synaptopathies Strategic Award, 165908) and National Institute for Health Research University College London Hospitals Biomedical Research Centre. Families 5–8 were collected as part of the SYNaPS Study Group collaboration funded by The Wellcome Trust and strategic award (Synaptopathies) funding. Research for these families was conducted as part of the Queen Square Genomics group at University College London, supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. NK is supported by intramural funds provided by King Faisal Specialist Hospital and Research Center, the National Plan for Science, Technology and Innovation program under King Abdulaziz City for Science and Technology and the King Salman Center for Disability Research. TVH is supported by an Erasmus University Rotterdam (EUR) fellowship. TSB’s lab is supported by the Netherlands Organisation for Scientific Research (ZonMW Veni, Grant 91617021), a NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation, an Erasmus MC Fellowship 2017 and Erasmus MC Human Disease Model Award 2018. TSB, IC and EA acknowledge support from COST action CA16118 that facilitated this collaboration.
Author information
Authors and Affiliations
Contributions
EP performed molecular biology experiments, with help from AN and DP. HvdL, WB and TvH performed zebrafish work. PvdB and EHJ performed enzymatic analyses. IC performed brain immunohistochemistry and supplied tissue samples. EA supplied tissue samples. MG generated iPSCs. WvI and WGdV performed and SY analyzed RNA-seq. SY performed gene homology search. Patient recruitment and diagnosis was performed in the different families as follows: Family 1: TSB, ASB, and EM phenotyped patient 1, MvS analyzed WES; Family 2: LB and MK phenotyped patients 2 and 3, KGM, AB, KR analyzed WES; Family 3: JNK and JB phenotyped patient 4, KGM, AB, KR analyzed WES. Family 4: AaF, FaM, RM and FaA phenotyped patient 5, EJK analyzed WES; Family 5: FZ and NR phenotyped patient 6, SE, HH analyzed WES; Family 6, family 7 and family 8: MM, AE, ZK, FMD, MD, EGK phenotyped patients 7–10, JV, RM, HH analyzed WES; Family 9: JH phenotyped patient 11, KKK, ABA analyzed WES; Family 10: MA, MAA, MAS, MA, RA, LAQ, WQ, SC, KA, MHAH, SA, KA, AD, FA, DC and NK phenotyped patients 12 and 13, performed WES analysis and PGD; Family 11: MDe, MYVM, MG, AGES and RM performed WES and phenotyped patients 14–17; Family 12: GRP phenotyped patient 19; Family 13: HAC phenotyped patient 20, KKK, ABA analyzed WES; Family 14, patient 21, and family 15, patient 22: KKK, ABA analyzed WES. RT, KR, KKK, PB, ABA, RM, HH provided genetic data for population analysis. TSB identified patient 1, conceived the study, obtained funding, supervised the lab work and wrote the manuscript, with input from all main authors. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
KGM, AB, RT and KR are employees of GeneDx, Inc. KR holds stock in OPKO Health, Inc. KKK, PB and ABA are employees of CENTOGENE AG.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Perenthaler, E., Nikoncuk, A., Yousefi, S. et al. Loss of UGP2 in brain leads to a severe epileptic encephalopathy, emphasizing that bi-allelic isoform-specific start-loss mutations of essential genes can cause genetic diseases. Acta Neuropathol 139, 415–442 (2020). https://doi.org/10.1007/s00401-019-02109-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-019-02109-6