Abstract
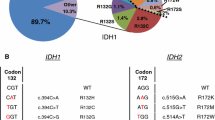
Diffusely infiltrating cerebral gliomas frequently carry point mutations in codon 132 of the isocitrate dehydrogenase 1 (IDH1) gene or in codon 172 of the IDH2 gene, which are both clinically important as diagnostic and prognostic markers. Here, we report on a method that allows for the rapid detection of IDH1 and IDH2 mutations based on pyrosequencing. The method is applicable to routinely processed tissue specimens and provides quantitative mutation data within less than one working day. Due to its high sensitivity, the technique may also be used for the diagnostic assessment of IDH1 or IDH2 mutation in tissue samples with low tumor cell content, such as the infiltration zone of diffuse gliomas. Using pyrosequencing and/or conventional cycle sequencing of IDH1 and IDH2 in 262 gliomas, we confirm frequent mutations in diffuse astrocytic and oligodendroglial gliomas, corroborate a prognostic role for IDH1 mutation in primary glioblastomas and show that pleomorphic xanthoastrocytomas generally lack mutations in these genes.




Similar content being viewed by others
References
Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A (2008) Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol 116:597–602
Bleeker FE, Lamba S, Leenstra S et al (2009) IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat 30:7–11
Capper D, Zentgraf H, Balss J, Hartmann C, von Deimling A (2009) Monoclonal antibody specific for IDH1 R132H mutation. Acta Neuropathol 118:599–601
Dang L, White DW, Gross S et al (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462:739–744
Dubbink HJ, Taal W, van Marion R et al (2009) IDH1 mutations in low-grade astrocytomas predict survival but not response to temozolomide. Neurology 73:1792–1795
Dufort S, Richard MJ, de Fraipont F (2009) Pyrosequencing method to detect KRAS mutation in formalin-fixed and paraffin-embedded tumor tissues. Anal Biochem 391:166–168
Felsberg J, Rapp M, Loeser S et al (2009) Prognostic significance of molecular markers and extent of resection in primary glioblastoma patients. Clin Cancer Res 15:6683–6693
Hartmann C, Meyer J, Balss J et al (2009) Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1, 010 diffuse gliomas. Acta Neuropathol 118:469–474
Ichimura K, Pearson DM, Kocialkowski S et al (2009) IDH1 mutations are present in the majority of common adult gliomas but are rare in primary glioblastomas. Neuro Oncol 11:341–347
Kang MR, Kim MS, Oh JE et al (2009) Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer 125:353–355
Kato Y, Jin G, Kuan CT, McLendon RE, Yan H, Bigner DD (2009) A monoclonal antibody IMab-1 specifically recognizes IDH1(R132H), the most common glioma-derived mutation. Biochem Biophys Res Commun 390:547–551
Kaulich K, Blaschke B, Numann A et al (2002) Genetic alterations commonly found in diffusely infiltrating cerebral gliomas are rare or absent in pleomorphic xanthoastrocytomas. J Neuropathol Exp Neurol 61:1092–1099
Korshunov A, Meyer J, Capper D et al (2009) Combined molecular analysis of BRAF and IDH1 distinguishes pilocytic astrocytoma from diffuse astrocytoma. Acta Neuropathol 118:401–405
Lof-Ohlin ZM, Nilsson TK (2009) Pyrosequencing assays to study promoter CpG site methylation of the O6-MGMT, hMLH1, p14ARF, p16INK4a, RASSF1A, and APC1A genes. Oncol Rep 21:721–729
Louis DN, Ohgaki H (2007) WHO classification of tumours of the central nervous system. IARC, Lyon
Mardis ER, Ding L, Dooling DJ et al (2009) Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 361:1058–1066
Mikeska T, Bock C, El-Maarri O et al (2007) Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis. J Mol Diagn 9:368–381
Mollemann M, Wolter M, Felsberg J, Collins VP, Reifenberger G (2005) Frequent promoter hypermethylation and low expression of the MGMT gene in oligodendroglial tumors. Int J Cancer 113:379–385
Ogino S, Kawasaki T, Brahmandam M et al (2005) Sensitive sequencing method for KRAS mutation detection by pyrosequencing. J Mol Diagn 7:413–421
Packham D, Ward RL, Ap Lin V, Hawkins NJ, Hitchins MP (2009) Implementation of novel pyrosequencing assays to screen for common mutations of BRAF and KRAS in a cohort of sporadic colorectal cancers. Diagn Mol Pathol 18:62–71
Parsons DW, Jones S, Zhang X et al (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321:1807–1812
Poehlmann A, Kuester D, Meyer F, Lippert H, Roessner A, Schneider-Stock R (2007) K-ras mutation detection in colorectal cancer using the pyrosequencing technique. Pathol Res Pract 203:489–497
Sanson M, Marie Y, Paris S et al (2009) Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol 27:4150–4154
Spittle C, Ward MR, Nathanson KL et al (2007) Application of a BRAF pyrosequencing assay for mutation detection and copy number analysis in malignant melanoma. J Mol Diagn 9:464–471
van den Boom J, Wolter M, Kuick R et al (2003) Characterization of gene expression profiles associated with glioma progression using oligonucleotide-based microarray analysis and real-time reverse transcription-polymerase chain reaction. Am J Pathol 163:1033–1043
Watanabe T, Nobusawa S, Kleihues P, Ohgaki H (2009) IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol 174:1149–1153
Watanabe T, Vital A, Nobusawa S, Kleihues P, Ohgaki H (2009) Selective acquisition of IDH1 R132C mutations in astrocytomas associated with Li-Fraumeni syndrome. Acta Neuropathol 117:653–656
Weber RG, Hoischen A, Ehrler M et al (2007) Frequent loss of chromosome 9, homozygous CDKN2A/p14(ARF)/CDKN2B deletion and low TSC1 mRNA expression in pleomorphic xanthoastrocytomas. Oncogene 26:1088–1097
Weller M, Felsberg J, Hartmann C et al (2009) Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the german glioma network. J Clin Oncol 27:5743–5750
Wick W, Hartmann C, Engel C et al (2009) NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J Clin Oncol 27:5874–5880
Yan H, Parsons DW, Jin G et al (2009) IDH1 and IDH2 mutations in gliomas. N Engl J Med 360:765–773
Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP (2004) A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res 32:e38
Zhang L, Kirchhoff T, Yee CJ, Offit K (2009) A rapid and reliable test for BRCA1 and BRCA2 founder mutation analysis in paraffin tissue using pyrosequencing. J Mol Diagn 11:176–181
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Location of the primer sequences used for IDH1 (A) and IDH2 (B) pyrosequencing. The PCR primer binding sites used in our analyses are labeled in grey (IDH1-For_Bio and IDH1-Rev for IDH1 amplification; IDH2-For and IDH2-Rev_Bio for IDH2 amplification). Location of the primers used for pyrosequencing are printed in blue and underlined (IDH1-PySeq and IDH2-PySeq, respectively). The commonly mutated codons 132 in IDH1 and 172 in IDH2 are printed in bold and labeled in yellow. In case of IDH1, the PCR primer binding sites used for amplification of a shorter amplicon of 72 bp with primers IDH1-For_Bio_short and IDH1-Rev_short are also indicated (red boxes). (DOC 19.5 kb)
Fig. S2
Comparison of IDH1 pyrosequencing results obtained on DNA extracted from frozen (cryo) or formalin-fixed paraffin-embedded (paraf) tissue samples of 10 different IDH1 mutant gliomas. Note that all mutations detected in high-molecular weight DNA extracted from frozen tumor tissue specimens were also detected in DNA samples obtained from paraffin material (values from corresponding sample pairs are indicated by the same color). Furthermore, the median percentages of mutant alleles detected by pyrosequencing (indicated by horizontal lines) were similar for both template sources, i.e., 39.5% (range: 18–43%) vs. 38.5% (range 30–46%). (TIFF 2.48 Mb)
Fig. S3
Sensitivity of IDH1 mutation detection by pyrosequencing. Shown are results of a dilution series using variable mixtures of IDH1 wild-type DNA and DNA from the tumor AA231, which carried a homozygous IDH1 mutation (c.395G>A; R132H) with a mutant allele percentage of 84% as determined by pyrosequencing. The ordinate indicates the mutant allele frequencies as detected by pyrosequencing. The abscissa shows the different concentrations of AA231 DNA in percent that were used as templates. The insert shows an enlarged part of the figure corresponding to the dilutions from 0 to 10% AA231 DNA. Note that pyrosequencing allows for a quantitative detection of mutant alleles and is already able to detect the IDH1 mutation when the template contained at least 4% of AA231 tumor DNA. To distinguish the presence of a mutation from background noise, a threshold of 5% mutant allele frequency (indicated by the horizontal line) is often used (4, 17) and recommended in the manual of the Pyromark Q24 machine (Qiagen). In our experiments, this 5% threshold was surpassed at a concentration of 8% of AA231 tumor DNA, corresponding to approximately 7% of mutant alleles. (TIFF 2.32 Mb)
Rights and permissions
About this article
Cite this article
Felsberg, J., Wolter, M., Seul, H. et al. Rapid and sensitive assessment of the IDH1 and IDH2 mutation status in cerebral gliomas based on DNA pyrosequencing. Acta Neuropathol 119, 501–507 (2010). https://doi.org/10.1007/s00401-010-0647-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-010-0647-4