Abstract
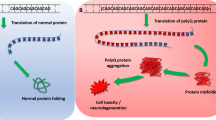
Spinocerebellar ataxia type 6 (SCA6) is an autosomal dominant neurodegenerative disease caused by a small polyglutamine (polyQ) expansion (control: 4–20Q; SCA6: 20–33Q) in the carboxyl(C)-terminal cytoplasmic domain of the α1A voltage-dependent calcium channel (Cav2.1). Although a 75–85-kDa Cav2.1 C-terminal fragment (CTF) is toxic in cultured cells, its existence in human brains and its role in SCA6 pathogenesis remains unknown. Here, we investigated whether the small polyQ expansion alters the expression pattern and intracellular distribution of Cav2.1 in human SCA6 brains. New antibodies against the Cav2.1 C-terminus were used in immunoblotting and immunohistochemistry. In the cerebella of six control individuals, the CTF was detected in sucrose- and SDS-soluble cytosolic fractions; in the cerebella of two SCA6 patients, it was additionally detected in SDS-insoluble cytosolic and sucrose-soluble nuclear fractions. In contrast, however, the CTF was not detected either in the nuclear fraction or in the SDS-insoluble cytosolic fraction of SCA6 extracerebellar tissues, indicating that the CTF being insoluble in the cytoplasm or mislocalized to the nucleus only in the SCA6 cerebellum. Immunohistochemistry revealed abundant aggregates in cell bodies and dendrites of SCA6 Purkinje cells (seven patients) but not in controls (n = 6). Recombinant CTF with a small polyQ expansion (rCTF-Q28) aggregated in cultured PC12 cells, but neither rCTF-Q13 (normal-length polyQ) nor full-length Cav2.1 with Q28 did. We conclude that SCA6 pathogenesis may be associated with the CTF, normally found in the cytoplasm, being aggregated in the cytoplasm and additionally distributed in the nucleus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Spinocerebellar ataxia type 6 (SCA6) is an autosomal dominant neurodegenerative disease caused by an expansion of a CAG repeat in exon 47 (the final exon) of the α1A voltage-dependent calcium channel (Cav2.1) (gene symbol: CACNA1A) [65]. SCA6 is allelic with three other dominantly inherited neurological diseases caused by different mutations in CACNA1A: episodic ataxia type 2, familial hemiplegic migraine [33], and progressive cerebellar ataxia [64]. Clinically, SCA6 patients show progressive cerebellar ataxia with an average age of onset at 45.5 years [17]. Besides progressive cerebellar ataxia, vertigo, oscillopsia, and episodic nature of cerebellar symptoms are characteristic features of SCA6 differentiating from other ataxias [8, 14, 19, 45]. Neuropathologically, degeneration is predominant in cerebellar Purkinje cells showing cerebellar cortical degeneration [8, 18]. In some cases, the inferior olivary complex may be affected [18, 44]. Thus, SCA6 is a prototype of cerebellar cortical degeneration, though degenerations outside the cerebello-olivary system may accompany [6]. SCA6 is found worldwide, with a relatively higher prevalence in Japanese [47].
SCA6 is considered one of at least nine known neurodegenerative disorders called polyglutamine (polyQ) diseases, which includes Huntington’s disease (HD), spinal and bulbar muscular atrophy (SBMA), SCA1, SCA2, Machado-Joseph disease/SCA3 (MJD), SCA7, SCA17, and dentatorubral-pallidoluysian atrophy (DRPLA) [60]. This is because the CAG repeat, which is transcribed into two different CACNA1A mRNA isoforms by an alternative splicing at the intron46/exon47 splice junction, is placed within a 3′-extended coding sequence of the major mRNA isoform, resulting in the CAG repeat being translated into a polyQ tract [1, 15, 65]. The CAG repeat in CACNA1A in normal individuals ranges from 4 to 20 repeats, whereas in SCA6 patients, this repeat is expanded usually ranging from 20 to 28 repeats [17, 45, 65], although longer expansions up to 33 repeats are rarely found [61]. Remarkably, CAG repeat/polyQ expansion in SCA6 is smaller than normal-length CAG repeats/polyQs in other polyQ diseases.
As the mutation of SCA6 is in CACNA1A encoding Cav2.1, a pore-forming subunit of P/Q-type voltage-dependent calcium channel essential for neurons [4, 30, 55], it is possible that such small polyQ expansion leads to neurodegeneration by functional alterations of Cav2.1 [5, 14, 21, 28, 35, 36, 50]. However, two recent studies on different SCA6 knock-in mice neither found that expanded polyQ affects the electrophysiological properties of Cav2.1 [37, 57], suggesting that the pathogenic mechanism of polyQ expansion in SCA6 is not merely due to functional changes of Cav2.1.
It has been known that Cav2.1 is highly expressed in cerebellar neurons and localizes primarily to nerve terminals, dendrites and Purkinje cell soma [59]. In SCA6, the Cav2.1 forms microscopic aggregates in Purkinje cells [15, 16]. Using a polyclonal antibody named A6RPT-C that recognizes the Cav2.1 carboxyl(C)-end, large rod-shaped aggregates were observed in cell bodies of SCA6 Purkinje cells [15]. Subsequent analysis using 1C2, a mouse monoclonal antibody that preferentially recognizes expanded polyQ tracts [51], also revealed the formation of granular aggregates [16]. However, the aggregates recognized by these antibodies did not completely co-localize [16], leaving the component(s) of the aggregates formed in SCA6 Purkinje cells obscure.
With regard to a toxicity of mutant protein, our group and others have shown that a 75–85-kDa C-terminal fragment of Cav2.1 (CTF), presumably generated by proteolytic cleavage of a recombinant full-length Cav2.1, was toxic in cultured cells, while full-length Cav2.1 was not [22, 24, 27]. This CTF was particularly toxic in cultured cells when it has an expanded polyQ [22, 27]. However, there is no direct evidence whether the CTF exists in human brains. Given that the CTF also exists in neurons and is toxic when having expanded polyQ, it would be particularly important to identify its area of expression in normal brains and how it is altered in SCA6. Moreover, it has not yet been clarified whether such a small polyQ expansion promotes aggregation of either full length Cav2.1 or any of its portions in SCA6 human brains. These fundamental questions remain unanswered since immunoblot analysis was not successful in human brains because of lack of sensitive antibodies against the Cav2.1.
In this study, we generate new antibodies (A6RPT-#5803 and 2D-1) against the C-terminus of Cav2.1 and demonstrate by immunoblot analysis that the CTF, which is expressed exclusively in the cytoplasmic soluble fraction of the human control cerebella, is aggregated in SCA6 brains harboring a small expansion (Q22 tract) in the Cav2.1. The CTF in SCA6 was also detected in the nuclear fraction, indicating that a small polyQ expansion affects intracellular location of CTF. A small polyQ expansion (Q28 tract), that is what seen in actual SCA6 patients, promoted recombinant CTF to aggregate and distribute in both the cell bodies and nuclei of cultured cells; however, it did not when it was expressed in the full-length Cav2.1. Considering CTF toxicity in cells, this study implies that the CTF is an important molecular component of SCA6 pathogenesis.
Materials and methods
The study was conducted in three parts: (1) development of new antibodies against the Cav2.1 C-region, (2) Western blot and immunohistochemical analysis of human control and SCA6 cerebella, and (3) investigation of recombinant, full-length Cav2.1 and CTF expressions in cultured cells.
Generation of plasmids for recombinant proteins
All recombinant human CACNA1A cDNAs (Fig. 1) were constructed from the previously described full-length CACNA1A clones [15]. Human genomic DNA samples were used to clone 11, 13, and 28 CAG repeats encoding Q11, Q13 (both normal), and Q28 (expanded), respectively. Here, an artificial clone with 165 CAG/CAA repeat encoding a 165-Q-stretch (extremely expanded; Q165) was also generated, as previously described [34]. Although the length of 165-Q-stretch is too long considering the actual number of CAG repeat expansion in SCA6 patients, it would allow us to see more dramatic effects of polyQ on the Cav2.1 protein aggregation.
Scheme representing recombinant (r) Cav2.1 fusion protein constructs. a Full-length rCav2.1 containing polyQ tracts of either Q11, Q28, or Q165. Epitope locations for different antibodies are also shown. b Recombinant CTF (rCTF), corresponding to the C-terminal 553 amino acids of Cav2.1. For both constructs, hemagglutinin A (HA) and c-Myc (Myc) are tagged in the N-term and C-term of Cav2.1/CTF, respectively. “AA” denotes amino acids in GenBank AB035726
Recombinant Cav2.1 CTF (rCTF) encoding the C-terminal 553 amino acids (AA) [AA#1954 to the C-end (AA#2506) in GenBank (http://www.ncbi.nlm.nih.gov/sites/entrez) Accession number #AB035727] was constructed by deducing the molecular mass size from the AA sequences. The present rCTF has longer AA sequences than that generated by Kordasiewicz et al. [22].
First, the full-length CACNA1A cDNA cloned in the SphI and XbaI sites of pcDNAI/Amp [15] was digested with BglII (at nucleotide number 5861 in AB035727) and XbaI, yielding a 1.7-kilobase (kb), 3′-fragment of the CACNA1A cDNA. This 1.7-kb fragment was cloned into pcDNA 3.1/myc-His (Invitrogen, Carlsbad, CA, USA). This clone was then subjected to site-directed mutagenesis to disrupt the original stop codon of CACNA1A mRNA and extend its coding sequence to include the C-tag, c-Myc being translated (named “Clone A”). Next, the 5′-region of this clone was amplified by PCR using forward primer 5′-AGTAAGGATCCGCCACCATGGCCTACCCATACGATGTTCCAG-3′, containing an artificial Bam HI site, the Kozak consensus (GCCACCATGG) sequence and a sequence encoding the N-tag hemagglutinin A(HA), and reverse primer 5′-TCTCCTTGAGGCCGCTTTCGTGAG-3′ to yield a 280-base-pair (bp) fragment from the 5′-region of Clone A. This PCR product was cloned into pCRII (Invitrogen). After checking its internal nucleotide (nt) sequence by DNA sequencing, this clone was digested with BamHI and BglII (corresponding to the nt number 5861 in AB035727), and the resultant fragment was cloned into the BamHI/BglII site of Clone A, producing the backbone construct for the recombinant HA-CTF-myc fusion protein. Four types of CAG repeats encoding Q11, Q13, Q28, and Q165 were cloned into this backbone construct using two KpnI sites flanking the CAG repeat sequence, producing the final rCTF-Q11, rCTF-Q13, rCTF-Q28, and rCTF-Q165 cDNA clones.
To construct the full-length recombinant (r) Cav2.1 cDNAs, we initially digested the full-length CACNA1A clones [15] with BglII, yielding a 6-kb DNA fragment containing the entire 5′-region of the full-length CACNA1A cDNA to the BglII site at nucleotide number 5861 in AB035726. This 6-kb DNA fragment was inserted into the unique BglII site of the rCTF cDNA-Q11, rCTF cDNA-Q28, and rCTF cDNA-Q165 clones to generate full-length rCav2.1 cDNA constructs encoding HA-full-length Cav2.1 (Q11, Q28, and Q165)-myc fusion proteins.
Antibody generation
In this study, two different anti-Cav2.1 antibodies (i.e., A6RPT-#5803 and 2D-1) were generated. For the rabbit polyclonal antibody A6RPT-#5803, the peptide “MERRVPGPARSESPRA” corresponding to the AA#2367–AA#2382 residues in the human Cav2.1 C-terminal region (GenBank #AB035726) was synthesized (see Fig. 1). The polypeptide exists as 41 AA downstream from the polyQ tract or 126 AAs upstream from the C-end (Fig. 1a). After being coupled to cystein, the synthetic peptide was injected into rabbits. Rabbit antisera were affinity purified, as previously described [15, 16]. Notably, A6RPT-#5803 specifically recognizes human Cav2.1 but not mouse, rat, or rabbit Cav2.1 (data available upon request). On the other hand, A6RPT-C which recognizes a polypeptide near the C-end [15] is expected to cross-react with the mouse and rat Cav2.1 because the antigenic peptide sequence is shared among the species [53]. Our recent antibodies on SCA6 knock-in mice using A6RPT-#5803 have demonstrated the presence of microscopic aggregates. A6RPT-#5803 is the same antibody “A6RPT-polyQ” described in Ref. [57].
The monoclonal antibody 2D-1, designed to detect the small peptide as A6RPT-C does, was produced by the conventional procedure [25]. In brief, BALB/c female mice (Sankyo Labo Service, Tokyo, Japan) were immunized with the synthetic peptide SRHGRRLPNGYYPAHGLAR, which corresponds to AA residues #2479–2497 near the C-end [15] (Fig. 1). Three days after the final injection, spleen cells of the immunized mice were fused with a myeloma cell line using 50% (w/w) polyethylene glycol (Roche Diagnostics, Basel, Switzerland). The fused cells were cultured on 96-well culture plates (Becton Dickinson Labware, Franklin Lakes, NJ, USA) in hypoxanthine aminopterin thymidine (HAT) selection medium (Invitrogen). Hybridoma cells producing objective antibodies were screened by enzyme-linked immunosorbent assay (ELISA) using 96-well plates coated with the synthetic peptides.
All the antibodies used here against Cav2.1 (i.e., A6RPT-#5803, 2D-1, and A6RPT-C) detect the Cav2.1 with an extended C-terminus encoded by the CACNA1A mRNAs with 3′-extended coding sequence due to the 5-nucleotide GGCAG insertion at the intron46/exon47 splice junction [15].
Cell culture, transfection, and immunoblot analysis of rCav2.1 proteins
PC12 cells, rat adrenal pheochromocytoma cell line, were cultured in Dulbecco’s Modified Eagle’s Medium containing 5% fetal bovine serum (FBS), 10% horse serum, and 1% penicillin/streptomycin in a humidified atmosphere of 5% CO2 at 37°C. PC12 cells were transfected with full-length rCav2.1 or rCTF cDNA constructs using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol.
Cells were harvested 48 h after transfection and then suspended in ice-cold MPER buffer (Pierce, Rockford, IL, USA) containing 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM phenylmethylsulfonyl fluoride (PMSF), and a protein inhibitor cocktail (Complete Mini tablets; Roche). Protein concentration was determined using the bicinchoninic acid assay (Pierce) according to the manufacturer’s protocol. Total extracts (50 μg) were suspended in a sodium dodecyl-sulfate (SDS) sample buffer [20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (pH 7.2), 120 mM NaCl, 10% glycerol, 2% SDS, 1 mM EDTA, 1 mM dithiothreitol (DTT), and a complete protease inhibitor cocktail] and separated by 12.5% polyacrylamide gel electrophoresis (PAGE) in a running buffer containing 1% SDS.
To detect the CTF in oligomer states, transfected cells were suspended for 20 min in an ice-cold, detergent-free buffer [25 mM Tris–HCl (pH 7.5), 140 mM NaCl, 1 mM EDTA and a complete protease inhibitor cocktail], and then centrifuged at 13,000g for 10 min at 4°C. The supernatant was diluted with a SDS sample buffer [62.5 mM Tris–HCl (pH 6.8), 2% SDS, 25% glycerol, and 0.01% bromophenol blue] for full-length rCav2.1, or with a different sample buffer without SDS [62.5 mM Tris–HCl (pH 6.8), 40% glycerol, and 0.01% bromophenol blue] for rCTF. After incubating at 37°C for 30 min, these samples were subjected to 12.5% PAGE in a running buffer containing 1% SDS to detect CTF dimers produced from the full-length rCav2.1, or to 7.5% PAGE under non-denaturing conditions to detect rCTF oligomers.
After PAGE, proteins were transferred onto a polyvinylidene fluoride membrane using 25 mM Tris-base, 192 mM glycine, and 10% methanol buffer. Blotted membranes were blocked for 1 h at room temperature with 5% skim milk in TBS-T [50 mM Tris (pH 7.5), 150 mM NaCl, and 0.05% Tween 20], washed three times in TBS-T, and then incubated with either of the following antibodies: A6PRT-#5803 (diluted with 5% skim milk in phosphate-buffered saline (PBS) at 1:5,000), A6PRT-C [15] (1:5,000), mouse monoclonal antibody 1C2 (1:10,000, Chemicon, Temecula, CA, USA), rabbit polyclonal anti-HA (sc805, 1:5,000, Santa Cruz Biotechnology, Santa Cruz, CA, USA) antibody or anti-Myc (sc40, 1:5,000, Santa Cruz) antibody. For loading controls, mouse monoclonal anti-calbindin-D28k antibody (CR955, 1:5,000, Sigma, St Louis, MO, USA), mouse monoclonal anti-β-tubulin antibody (1:5,000, Becton Dickinson Bioscience, San Jose, CA, USA), and rabbit polyclonal anti-Histone H3 antibody (1:5,000, Cell Signaling, Boston, MA, USA) were used. Primary antibodies were detected with the appropriate secondary anti-mouse or anti-rabbit IgG labeled with peroxidase (1:5,000, Jackson ImmunoResearch, Baltimore, PA, USA), and finally visualized using ECL (GE Healthercare, UK). To check the specificities of A6RPT-#5803 and 2D-1, immunoblot procedures were undertaken in a special device (ATTO, Tokyo, Japan) that makes it possible to apply different antibodies to one blotted membrane at a time.
Brain tissues
Human tissues from six controls and two SCA6 brains (Table 1) were analyzed by immunoblot analysis. All brains were obtained at autopsy performed under the families’ written consents. The study was approved by the institutional review board of ethics of our institutions and conformed to the tenets of the Declaration of Helsinki. The obtained brains were immediately frozen and stored at −80°C until use. Tissue blocks from the frozen cerebella, occipital cortex, and thalamus were used for immunoblot analysis. The number of CAG repeats in CACNA1A was also examined in each subject, as previously described [17]. The results showed that all SCA6 patients coincidentally harbored the same 22 CAG repeats in their expanded alleles. Note that these expanded polyQs (Q22) in the SCA6 patients are only 15Q tract longer than the shortest polyQ (Q7), or 5Q tract longer than the longest normal-polyQ (Q17) in the control group (Table 1).
Immunoblot analysis of human brain tissue samples
All protein extractions were carried out in a cold room maintained at 4°C, unless specifically described. Human cerebellar cortices (0.1 g) were homogenized in radio-immunoprecipitation assay (RIPA) buffer [50 mM Tris–HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA (ethylene glycol tetraacetic acid), 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100, 1% PMSF, and the complete protease inhibitors] with the PHYSCOTRON handy micro homogenizer (Microtec Nition, Japan). Then, the homogenate was sonicated with a microprobe for 30 s and centrifuged at 13,000g for 20 min. The supernatants (crude extracts) were collected and stored at −80°C.
To clarify the Cav2.1 expression pattern in more detail, subcellular fractionations were performed at 4°C, as previously described [40] (Fig. 2). Brain samples (0.5 g each of either, cerebellum, occipital cortex, or thalamus) were homogenized in ten volumes of buffer containing 0.32 M sucrose, 20 mM HEPES homogenization buffer (pH 7.2), 1 mM DTT, 1 mM PMSF, and a complete protease inhibitor cocktail, using ten strokes of tight-fitting glass homogenizers. The homogenate was centrifuged at 1,000g for 15 min, and the resulting supernatant (fraction “C”) was transferred to a new tube for further analysis of the cytosolic and membrane fraction. The pellet was rinsed four times with ice-cold homogenization buffer and then homogenized, diluted to 1.2 ml with 0.25 M sucrose/buffer TKM [50 mM Tris–HCl (pH 7.5), 25 mM KCl, and 5 mM MgCl2], and mixed with two volumes of 2.3 M sucrose/buffer TKM. The mixture was then layered on top of the 0.6 ml of 2.3 M sucrose/buffer TKM in a SW41 centrifuge tube (Beckman, CA, USA) and centrifuged at 10,000g for 1 h. The pellet was re-suspended in 1000 μl of 0.25 M sucrose/buffer TKM and re-centrifuged at 2,000g for 10 min. The resultant pellet was re-suspended in 0.25 M sucrose/buffer TKM as the final nuclear fraction (N) stored at −80°C until analysis.
Protocol for detailed investigation of Cav2.1 in human brain (see also “Materials and methods”). In step 1, the brain tissue was fractionated to separately enrich cytosolic (C) and nuclear (N) proteins in a sucrose buffer. In step 2, the resultant cytosolic fraction (C) was centrifuged to generate a sucrose-soluble fraction (C1) and a pellet (P1). In step 3, the P1 fraction was dissolved using a buffer containing SDS and then centrifuged to produce a supernatant containing SDS-soluble protein fractions (C2) and a pellet of SDS-insoluble proteins (P2). In step 4, the P2 fraction was dissolved using formic acid (FA) (C3). Thereafter, the four types of extracts (N, C1, C2, and C3) were separated by SDS-PAGE
The fraction C was further processed by centrifugation at 16,000g at 4°C for 5 min, as previously described with slight modifications [10, 26]. The supernatant was collected as a sucrose-soluble cytosolic fraction (C1). The resultant pellet was re-suspended in 100 μl of SDS buffer [20 mM HEPES (pH 7.2), 120 mM NaCl, 10% glycerol, 2% SDS, 1 mM EDTA, 1 mM DTT, and a protease inhibitor cocktail] at 37°C for 30 min, and then separated by centrifugation at 16,000g at room temperature. The supernatant was collected as a SDS-soluble cytosolic fraction (C2). The SDS-insoluble pellet was re-suspended in 100% formic acid (FA), incubated at 37°C for 1 h, and then dried by centrifugation in vacuo. The resulting dried material was re-suspended in a sample buffer [62.5 mM Tris–HCl (pH 6.8), 2% SDS, 25% glycerol, and 0.01% bromophenol blue] and neutralized with 1 M Tris–HCl, yielding a SDS-insoluble/FA-soluble cytosolic fraction (C3). The rest of the procedures for measuring protein concentration and PAGE (using 12.5% acrylamide gels or 5–20% acrylamide gradient gels) were performed similarly as described in “Cell culture, transfection, and immunoblot analysis of rCav2.1 proteins”.
Immunohistochemistry of human brain tissues
Six controls and seven SCA6 brains were studied (Table 1). Four of these seven SCA6 brains have already been described [16, 18, 39]. Brains were obtained as described earlier and fixed in 10% neutral-buffered formalin. Tissue samples were paraffin-embedded and prepared for neuropathological examination.
Immunohistochemistry was performed as previously described [15, 16]. Cerebellar tissue blocks containing the cerebellar cortex and deep cerebellar nuclei, brainstem (midbrain, pons, and medulla), cerebrum (frontal, temporal and occipital lobes, the striatum, and the thalamus), and spinal cord (cervical spine) were selected and sectioned at 6-μm thickness. For antigen retrieval, sections were deparaffinized with xylene, washed with distilled water, and boiled for 1 min three times in 10 mM citrate buffer (pH 7.4). For 1C2 immunohistochemistry, sections were additionally immersed in FA for 5 min. After antigen retrievals, all sections were washed in distilled water, treated with 0.3% (v/v) hydrogen peroxide in methanol to quench endogenous peroxide, and then incubated with a mixture of normal goat and horse sera for 20 min. Finally, sections were incubated overnight at 4°C with anti-Cav2.1 antibodies (i.e., A6RPT-C, 2D-1, and A6RPT-#5803, all at 1:500) or with 1C2 (1:4,000). The primary antibodies were serially detected with the appropriate biotinylated anti-rabbit or anti-mouse IgG (Vector, CA, USA), avidin-biotinylated-peroxidase complex (Vector), and finally developed with 3,3′-diaminobenzidine and hydrogen peroxide as chromogen. Between each step, sections were washed with PBS containing 0.1% Triton-X (PBS-T) three times. Staining specificity was assessed for each primary antibody both by substituting the primary antibody with rabbit or mouse IgG solution (Vector) and by absorption tests as previously described [15].
For double immunofluorescent labeling, sections were similarly treated and incubated with primary antibodies overnight. After washing with PBS, primary antibodies were detected with Texas-Red conjugated anti-rabbit goat IgG and fluorescence isothiocyanate conjugated anti-mouse horse IgG (Vector; both diluted at 1: 1,000 with PBS) for 1 h at room temperature. Sections were then washed with PBS-T and examined under a confocal light microscope (TCS-NT, Leica, Germany, LSM 510META, Carl Zeiss).
Immunocytochemistry of Cav2.1 in cultured PC12 cells
PC12 cells expressing full-length rCav2.1 or rCTF were fixed with 4% paraformaldehyde in PBS for 15 min at room temperature and washed three times in PBS. After treatment with 0.4% TritonX-100 in PBS for 15 min, cells were incubated with in a blocking solution (PBS containing 5% bovine serum albumin and 0.1% Tween 20) for 30 min. Cells were incubated with the anti-tag antibodies, A6RPT-#5803 and 1C2 for 1 h and then incubated with fluorescein-labeled anti-mouse IgG (H + L) (1:1,000; Vector Laboratories) and Cy3-conjugated affinity-purified anti-rabbit IgG (H + L) (1:1,000; Rockland, PA, USA) for 1 h. Slides were visualized by confocal microscope (LSM 510META, Carl Zeiss).
Quantitative image analysis
Images were analyzed using WinRoof software ver 5.7 (Mitani Corporation, Japan). For measurements of nuclear and cytoplasmic fluorescence, nuclear and whole cell regions of interest (ROIs) were generated by density slicing the Hoechst and A6RPT-#5803 images, and cytoplasmic ROIs were obtained by subtraction. Then, intensity of the fluorescence emitted from Cy3-conjugated anti-rabbit IgG antibody detecting the primary A6RPT-#5803 antibody was measured in each ROIs by mean intensity of area. Intensity of untransfected cells was utilized to determine the threshold and calibration.
Fluorescence measurements were performed in 300 cultured cells (n = 100 per condition). Three groups were statistically analyzed using one-way ANOVA. Then, differences between two groups (e.g., rCTF-Q13 vs. rCTF-Q28) were analyzed using both Tukey–Kramer and Bonferroni/Dunn tests. Results were plotted as mean ± SD, and differences were considered significant at P < 0.05.
Results
Generation and characterization of new anti-human Cav2.1 antibodies A6RPT-#5803 and 2D-1
The polyQ tract lies in the cytoplasmic domain of extended C-terminal region of Cav2.1 [15, 65]. Thus, antibodies reacting with the C-terminus are important for exploring SCA6 pathogenesis. Specificities of two new antibodies against this extended C-terminal region, A6RPT-#5803 (rabbit polyclonal) and 2D-1 (mouse monoclonal), were tested by performing immunoblot analysis on protein extracts from PC12 cells expressing the cDNA construct encoding the rCTF-Q28. Both A6RPT-#5803 and 2D-1 faithfully detected the rCTF-Q28 with an approximate molecular size of 75–85 kDa (Fig. 3).
Specificities of new anti-Cav2.1 antibodies A6RPT-#5803 (a) and 2D-1 (b). A special device was used to incubate one blotted membrane with different antibodies in different lanes at once. rCTF-Q28 expressed in PC12 cells was probed with different antibodies. A6RPT-#5803 (Membrane A) and 2D-1 (Membrane B) faithfully recognized rCTF-Q28, as recognized by anti-HA antibody, anti-Myc antibody, anti-polyQ antibody 1C2, and A6RPT-C
The CTF generated in cultured cells expressing full-length rCav2.1
Next, the full-length rCav2.1 proteins containing either one of the three different polyQs (i.e., Q11, Q28, and Q165) were transiently expressed in cultured PC12 cells. When their protein extracts were analyzed by immunoblot analysis using A6RPT-#5803, we observed strong bands at about 250 kDa (Fig. 4a, arrow), the expected molecular mass size of the full-length Cav2.1s [15, 33, 65]. Notably, reactions were also detected at the stacking gels for full-length rCav2.1 with Q165 (Fig. 4a, asterisk) and that with Q28, but were very weak for rCav2.1 with Q11. This indicates that proteins can become insoluble when the polyQ tract is expanded. In addition to full-length rCav2.1s, clear bands at molecular masses ranging between 75 and 110 kDa were also demonstrated (Fig. 4a, arrowheads). Differences in sizes of these smaller fragments corresponded to the differences in their polyQ lengths. Although the antibody against C-tag (anti-Myc antibody) detected both proteins at 250 and 75–110 kDa (Fig. 4b), the antibody against N-tag (anti-HA antibody) failed to detect the 75–110-kDa protein (Fig. 4c). Hence, the 75–110-kDa protein detected by A6RPT-#5803 should correspond to the CTF of Cav2.1. With the anti-expanded polyQ antibody 1C2, the CTFs generated from the full-length rCav2.1(Q165) and full-length rCav2.1(Q28) (arrowheads) as well as the full-length rCav2.1(Q165) (arrow) were clearly detected (Fig. 4d). Detection of rCTF-Q28 by 1C2 was not expected, since it has been believed that length larger than Q37 is needed for detection by 1C2 [51].
The CTF of Cav2.1 is generated in PC12 cells expressing full-length rCav2.1 fusion proteins. a Immunoblot analysis of protein extracts from PC12 cells expressing full-length rCav2.1 containing either Q11, Q28, or Q165. The blot was probed with A6RPT-#5803. The arrow indicates full-length rCav2.1 at about 250 kDa, and the arrowheads denote the C-terminal fragment with different sizes depending on the polyQ length. The asterisk indicates aggregated proteins in the stacking gel seen in full-length rCav2.1 with Q28 and Q165. “Unt” denotes protein extracts from untransfected cells as a negative control. b The anti-Myc antibody, detecting the C-end tag in the full-length rCav2.1 fusion protein, recognized both 250 and 75–110 kDa proteins. c The anti-HA antibody, directed at the N-terminus of the fusion protein, recognizes only the full-length proteins, suggesting that the 75–110-kDa bands are the CTF. d The 1C2 antibody recognized full-length rCav2.1 with Q165, two CTFs from Q165 and Q28 full-length rCav2.1 constructs but not obviously recognize rCav2.1s (full-length and CTF) with Q11 or full-length rCav2.1 with Q28
The CTF is present in human cerebella
Next, we examined whether the CTF exists in human brains. First, we analyzed crude cerebellar extracts with A6RPT-#5803. Consistent with the results from cultured PC12 cells expressing full-length rCav2.1s, we observed full-length Cav2.1 at 250 kDa (Fig. 5a, arrow) and intense bands at 75–85 kDa (Fig. 5a, arrowhead) in both control and SCA6 brains. Two additional bands (approximately 110 and 160 kDa) were also seen in both control and SCA6 brains, with the 160-kDa protein showing stronger reactivity than the 110-kDa protein. Because these proteins were not observed from the extracts of full-length rCav2.1 protein, they could be either the products processed from the full-length Cav2.1, products made by autolysis of the full-length Cav2.1, or the Cav2.1 isoforms generated by alternative splicing, as previously described for rat Cav2.1 [38].
The CTF, normally found exclusively in the cytosolic fraction, is also detected in the nuclear fraction of human SCA6 cerebella. a Immunoblot analysis of crude extracts of human cerebella detected by A6RPT-#5803 (controls in lanes 1–4 and SCA6-1 in lane 5). The arrow indicates the full-length Cav2.1 at about 250 kDa, and the arrowhead indicates the protein possibly corresponding to the 75–85-kDa CTF. The blots were probed with antibodies to β-tubulin (middle), and calbindin (bottom) as loading controls. b Immunoblot analysis of sucrose-soluble cytosolic (C1) and nuclear (N) fractions obtained from control cerebella (controls 1–5). A6RPT-#5803 detected both 75–85-kDa CTF and 150–200-kDa protein only in the cytosolic fractions, whereas A6RPT-C detected only 75–85-kDa CTF. Anti-β-tubulin and anti-Histon3 antibodies were used as fractionation control of cytosolic and nuclear proteins, respectively. c Immunoblot analysis of SCA6 and control brains probed by A6RPT-#5803 or 1C2. In the SCA6 brain (SCA6-1: patient 3 in Table 1), the 75–85-kDa CTF was present both in the sucrose-soluble cytosolic (C1) (arrow) and nuclear (N) (arrowhead) fractions. The CTF in the nuclear fraction was also detected by 1C2 (arrowhead). Note that this difference is based on only a small polyQ expansion (<17 polyQ difference) in Cav2.1. d Immunoblot analysis on the cerebella of a different SCA6 patient (SCA6-2: patient 4) and MJD patient. The CTF was not detected in the nuclear fraction of the cerebellum of the MJD patient but was detected in the cerebellum of the SCA6 patient (SCA6-2) (arrowhead). e The CTF is seen in the cytosolic (C) sucrose-soluble fraction in both control and SCA6 brains (Ctx the occipital cortex; Bg the thalamus). However, the CTF is not seen in any nuclear fractions from control SCA6 brains
The CTF with expanded polyQ tract present in the nuclear extract of SCA6 cerebella
The intracellular location of Cav2.1 in human brains, particularly of the CTF, cannot be addressed in immunoblotting on crude homogenates. To overcome this problem, we performed subcellular fractionation using a four-step protocol according to the solubility against sucrose, SDS, and FA (Fig. 2). In a sucrose-soluble cytosolic fraction (C1) of a control cerebellar cortex probed with A6RPT-#5803, we observed two clear bands with molecular masses ranging between 75–85 and 150–200 kDa, conceivably corresponding to the CTF and Cav2.1 isoforms, respectively (Fig. 5b). No obvious bands were observed in the sucrose-soluble nuclear fraction (N) from control cerebella. A6RPT-C also detected the 75–85-kDa protein in the C1 but not in the N, whereas it did not detect the 150–200-kDa protein. Since A6RPT-C and A6RPT-#5803 both detected the same 75–85-kDa protein fragment, we considered this fragment as the CTF in the human brain. On the other hand, the 150–200-kDa protein was not recognized by A6RPT-C. These results may suggest that A6RPT-C has higher specificity, but less sensitivity, to the CTF than A6RPT-#5803.
In the SCA6 brains, A6RPT-#5803 detected the 75–85-kDa CTF not only in C1 (Fig. 5c, arrow) but also in the sucrose-soluble nuclear fraction (N) (arrowhead). The 150-kDa protein fragment seen in C1 was not detected in N. When we probed with 1C2, the CTF in the N of SCA6 cerebella was detected (Fig. 5c, arrowhead). To further determine whether the nuclear CTF localization was specific to SCA6, we performed similar experiments on protein extracts from a MJD cerebellar cortex using A6RPT-#5803. Results showed that the CTF in a MJD cerebellum was observed only in the cytosolic fraction (C1) as in other control brains (Fig. 5d). This indicates that nuclear CTF localization is specific to SCA6 and is not merely a result of cerebellar degeneration.
We next examined the occipital cortex and the thalamus to see whether findings in the SCA6 cerebella are also seen in other regions of SCA6 brains. We found that the 75-kDa CTF is present in the sucrose-soluble cytosolic fractions (C1) of both controls and SCA6 extracerebellar tissues (Fig. 5e). However, the CTF was not detected in their nuclear fractions (N), indicating that the presence of CTF in nuclear fraction is restricted only in the SCA6 cerebellum. We also noted that the level of expression could be relatively low in these two regions compared to the cerebellum. This is in accord with previous observation that Cav2.1 is most abundant in the cerebellum [15].
Detection of insoluble Cav2.1 protein fragments in the cytosolic fraction specific to SCA6
Sucrose-insoluble cytosolic proteins were dissolved in SDS (C2 in Fig. 2), and the resultant SDS-insoluble materials were further dissolved in FA (C3). Considering that Cav2.1 is a membrane-bound protein, it is conceivable that the protein extracted by SDS represents the basic nature of Cav2.1 in the human brain.
In both control and SCA6 cerebella, we observed four major bands with approximate molecular sizes of 25, 37, 85, and 170 kDa in SDS-soluble extracts (C2) (Fig. 6a). A band of approximately 85 kDa would correspond to the CTF (arrowhead), whereas bands at 25 and 37 kDa may be processed fragments of Cav2.1. A band above 250 kDa (arrow), more clearly seen in SCA6 cerebella, could be the full-length Cav2.1. Smears above 100 kDa (filled star) were also seen more clearly in SCA6 cerebellar extracts (Fig. 6a).
Cytoplasmic aggregates in SCA6 brains contain the Cav2.1 CTF. a Immunoblot analysis using A6RPT-#5803. In the SDS-soluble cytosolic fractions (C2), four major bands with approximate molecular sizes of 25, 37, 85, and 170 kDa are seen both in control and two SCA6 cerebella. Bands at 25 and 37 kDa may correspond to the processed fragments of Cav2.1. A band at approximately 85 kDa indicates the CTF. A band above 250 kDa (arrow), more clearly seen in SCA6 cerebella, is considered the full-length Cav2.1. Smears above 100 kDa (filled star) are also seen more clearly in SCA6 cerebellar extracts. In SDS-insoluble/formic acid (FA)-soluble extracts (C3), smears are seen above 50 kDa exclusively from SCA6 cerebella (open star). b In short exposure film of the FA-soluble C3 fractions, a band in between 75 and 100 kDa corresponding to the CTF is seen (arrowheads). c Immunoblot analysis using 1C2. Both extracts (C2 and C3) from SCA6 cerebella contain firm aggregates attached on top of the gel, particularly in the FA-soluble C3 fraction but not in control cerebella. d Protein reactions are seen in the SDS-soluble cytosolic fraction (C2) of both control and SCA6 brains. However, reactions are not seen in any SDS-insoluble/FA-soluble fractions (C3) of these brains (Ctx the occipital cortex; Bg the thalamus)
In FA-soluble extracts (C3), smears (open star) were observed above 50 kDa exclusively from SCA6 cerebella (Fig. 6a). On shorter exposure film, the CTF was revealed in this smear (Fig. 6b, arrowhead), indicating that the CTF would be one of the important constituents of aggregates. There was no obvious band corresponding to the full-length Cav2.1 in this C3 fraction.
When we probed SDS-soluble (C2) and FA-soluble (C3) cytosolic fractions with 1C2, immunoreactions were found exclusively in the SCA6 cerebellar extracts (Fig. 6c). Notably, both C2 and C3 fractions contained firm aggregates attached on top of the gel, particularly in the C3 fraction (Fig. 6c, arrowhead). These data indicate that protein components in human SCA6 cerebella have tendencies for aggregation. The 75-kDa protein detected with A6RPT-#5803 was not recognized with 1C2, while the high molecular weight band at the stacking gel was clearly demonstrated only by 1C2. These differences may be partly due to differences in sensitivities and specificities of these two antibodies: A6RPT-#5803 specifically detects the C-portion of Cav2.1 protein, whereas 1C2 detects expanded polyQ tract longer than that causes SCA6. 1C2 may be probably more sensitive to conformational changes promoted by polyQ stretch, or 1C2 may cause multiple bivalent bindings of the antibody on the homopolymeric epitope than A6RPT-#5803 does [51].
The 75–85-kDa CTF in the SDS-soluble cytosolic fractions (C2) was seen in the occipital cortex C2 fractions of control and SCA6 brains, while it was not evident in the thalamus from the same brains (Fig. 6d). This seemed to be due to the difference in the level of Cav2.1 expression; the thalamus showed lower expression level compared to the cerebral cortex [15]. Smear formation above 100 kDa, seen in the same fraction of SCA6 cerebella, was not evident either in the occipital cortex or the thalamus of the same SCA6 brain. In the SDS-insoluble/FA-soluble (C3) cytosolic fractions of the two extracerebellar tissues no obvious bands were detected (Fig. 6d), again demonstrating that the CTF and smear in C3 fractions was restricted in the SCA6 cerebella (Fig. 6a, open star). It is possible that the smear seen in the C2 and C3 fractions of SCA6 cerebella (Fig. 6a, filled and open stars) are associated with the Cav2.1 aggregation processes, though the difference in the Cav2.1 expression levels should also be considered.
Microscopic aggregates formed in the cytoplasm of SCA6 Purkinje cells
Immunohistochemistry of control cerebella using A6RPT-#5803 showed a weak and diffuse immunoreactivity in the cell bodies and dendrites of Purkinje cells (Fig. 7a). Although a previous study suggested that the C-terminal region of Cav2.1 is expressed in the nuclei [22], no obvious immunoreactivity was observed in the nuclei of any neurons in our experiment. Our finding of predominant cytoplasmic expression of Cav2.1 is consistent with our finding on immunoblotting (Fig. 5b), and is further supported by previous immunohistochemical studies on humans [15] and other species [43, 48, 59]. Other neurons demonstrated very weak immunoreactivity in their cell bodies (data not shown).
Immunohistochemistry for Cav2.1 extended C-terminal region in control and SCA6 cerebella. a A Purkinje cell in control cerebellum shows only a weak and diffuse immunoreactivity against A6RPT-#5803. Note that the nuclei of Purkinje or granule cells are not obviously stained (Control 1). b In SCA6 cerebella (patient 3), microscopic aggregates are abundantly seen with A6RPT-#5803. Most aggregates are small granules present in the cell body (black arrowhead) and dendrites (red arrow). Also, clusters of granules (white arrowhead) or a large rod-shaped aggregate (black arrow) is seen. c On higher magnification, abundant small granules immunoreactive for A6RPT-#5803 are seen (patient 3). Their sizes ranged approximately from 0.5 to 5 μm. Note that many aggregates are present in the perinuclear cytoplasm and dendrites in this neuron, but an aggregate also appears to locate within the nucleus (arrow). d A large rod-shaped aggregate is seen with 2D-1 in a SCA6 Purkinje cell (patient 1). e Large cytoplasmic aggregates are demonstrated using A6RPT-C (arrows) (patient 3). f Aggregates recognized by A6RPT-#5803 disappear with pre-absorption test. g Aggregates recognized by 2D-1 also disappear with pre-absorption test. h–k Cytoplasmic aggregates are not seen either in the pyramidal neuron in the occipital cortex (h A6RPT-#5803; i 1C2) or in the thalamus (j A6RPT-#5803; k 1C2) (patient 4) (scale bars a, b, d–h 50 μm; c, i–l 25 μm)
In SCA6 brains, small granular aggregates were abundantly seen in the cell bodies of Purkinje cells (Fig. 7b, c), demonstrating a distinct difference from controls. The frequency of these cytoplasmic aggregates was very high; hence, it was observed in nearly all Purkinje cells remaining in SCA6 cerebellar cortices.
Microscopic aggregates detected by A6RPT-#5803 in SCA6 Purkinje cells most commonly appeared as fine cytoplasmic granules with diameters ranging approximately from 0.5 to 5 μm (Fig. 7c). However, they sometimes existed in clusters (Fig. 7b, white arrowheads) or appeared as rods (Fig. 7b, black arrow). While aggregates were usually seen in the cell body or proximal dendrites, they were found around the nuclei (peri-nuclear), or even appeared to exist within the nuclei under a light microscopic observation (Fig. 7c, arrow). Diffuse labeling of the nucleus, as described in DRPLA, MJD, and other polyQ diseases [63], was never observed. The nuclei of Purkinje cells were not obviously stained as in controls.
Staining with 2D-1 also showed cytoplasmic aggregates (Fig. 7d). While A6RPT-#5803 picked up both granular and large rod-shaped aggregates, A6RPT-C detected only large aggregates (Fig. 7e), as described previously [15, 16]. Pre-absorption tests for A6RPT-#5803 and 2D-1 confirmed the immunohistochemical detection of these aggregates (Fig. 7f, g).
Similar aggregates were not confirmed by either A6RPT-#5803 or 1C2 in any other neurons, including granule cells and dentate neurons in the cerebellum, inferior olivary neurons in the medulla oblongata, pyramidal neurons in the cerebral cortex, or neurons in the basal ganglia (Fig. 7h–k). These results led us to conclude that aggregates formations are restricted in the SCA6 Purkinje cell.
Double labeling immunofluorescence studies with A6RPT-#5803 and 1C2 showed nearly complete overlap; “granular” aggregates in the cell body of SCA6 Purkinje cells were detected by both antibodies (Fig. 8). The only exception for this was seen in large aggregates (white arrow) detected more obviously by A6RPT-#5803. As previously reported [16], large aggregates recognized by A6RPT-C were also not labeled by 1C2. Taking these facts into account, A6RPT-#5803 detects both aggregates recognized by A6RPT-C and 1C2.
Double immunofluorescent confocal images of the Cav2.1 extended C-terminus (A6RPT-#5803) and expanded polyQ (1C2) in a SCA6 Purkinje cell (patient 3). Most granular aggregates recognized by A6RPT-#5803 (a red) are also recognized by 1C2 (b green) (c merged: yellow). A cluster of granular aggregates detected by A6RPT-#5803 is very weakly recognized by 1C2 (white arrow)
Oligomers and visible aggregates formed by CTF in cultured cells
Immunoblot analysis using A6RPT-#5803 in SCA6 cerebella suggested that the endogenous CTF with a small polyQ expansion has a tendency to aggregate. To further confirm this potential to aggregate, we performed immunoblot analysis with A6RPT-#5803 under mildly denaturing conditions. First, rCTF with three different polyQs (Q13, Q28, and Q165) was expressed in PC12 cells. When the protein extracts from these cells were boiled in the presence of SDS, clear bands were seen with different sizes corresponding rCTFs (Fig. 9a). However, when proteins were extracted without any detergents and separated in non-denaturing (native) polyacrylamide gel, we found intense and broad bands corresponding to be heterogeneous CTF proteins, namely, “CTF oligomers” (Fig. 9b, open arrowheads). These observations support the idea that the rCTF could have a tendency to form oligomers. Next, to address whether native CTF cleaved from full-length rCav2.1 has a potential to form oligomers, we treated PC12 cells expressing full-length rCav2.1 proteins for 48 h with detergent-free buffer, and then incubated under a mild condition with 2% SDS sample buffer for 30 min at 37°C just before PAGE. We observed immunoreactive bands at 150–200 kDa that appeared to correspond to the CTF dimers (Fig. 9c, open arrowhead). This would indicate that the CTF with even a small polyQ indeed has a potential to aggregate.
Detection of CTF dimer/oligomer from PC12 cells expressing rCav2.1s. a Immunoblot analysis of rCTF with A6RPT-#5803 in a denaturing condition. A6RPT-#5803 recognizes rCTF monomers. b Immunoblot analysis of rCTF with A6RPT-#5803 in a non-denaturing condition. By native-gel PAGE, broad bands (open arrowheads) presumably corresponding to heterogeneous CTF proteins are seen besides clear bands corresponding to the CTF monomers (filled arrowheads). c Immunoblot analysis of full-length rCav2.1 in a mildly denaturing condition using A6RPT-#5803 (see “Materials and methods” for details). Weak immunoreactive bands (open arrowhead) are presumed to reflect CTF dimers since their molecular sizes are double of the CTF monomers (filled arrowhead)
To clarify whether the intracellular distribution of CTF differs from that of full-length Cav2.1, we performed immunocytochemistry on PC12 cells, in which either full-length rCav2.1 or the rCTF was transiently expressed. The full-length rCav2.1 generally took perinuclear cytoplasmic distribution (Fig. 10a). There were no obvious 1C2-positive aggregates in PC12 cells expressing full-length rCav2.1 with normal (Q11) and expanded (Q28) polyQs. 1C2-positive aggregates were observed only in cells expressing the full-length rCav2.1 with Q165 (arrow). When rCTFs were expressing, rCTFs-Q13 did not show no obvious aggregates in the PC12 cells (Fig. 10b). However, 1C2-positive aggregates were found in cells expressing rCTF-Q28 or rCTF-Q165 (Fig. 10b). These microscopic aggregates were present mainly in the cytoplasm of PC12 cells (arrowheads) but were also seen in their nuclei (arrows).
rCTF with expanded polyQ forms visible aggregates with increased nuclear distribution in cultured PC12 cells. a Intracellular distribution of full-length rCav2.1 examined by immunocytochemistry using A6RPT-#5803 (red) and 1C2 (green) antibodies. The nuclei were visualized with Hoechst (blue). Full-length rCav2.1 proteins (Q11, Q28, and Q165) localize in the cytoplasm and only the full-length rCav2.1 with Q165 forms 1C2-positive aggregates (arrow) (scale bars 10 μm). b Intracellular rCTF distribution. rCTFs with different polyQs (Q13, Q28, and Q165) were stained with 1C2 (green) or A6RPT-#5803 (red). The CTF with expanded polyQ (Q28 and Q165) formed visible aggregates in both the cytoplasm and nucleus but predominantly in the cytoplasm, whereas rCTF-Q13 did not. Arrows indicate cytoplasmic aggregates and arrowheads nuclear aggregates (scale bars 10 μm). c Histogram showing ratios of nuclear to cytoplasmic fluorescence detecting A6RPT-#5803 immunoreactivities in PC12 cells transfected with CTF-polyQ13 (red), Q28 (green), and Q165 (blue) (n = 100 per condition). rCTF distribution in the nucleus increased in a polyQ length-dependent fashion. d Mean A6RPT-#5803 cell body fluorescence in PC12 cells transfected with rCTF-polyQ (Q13, Q28, and Q165) (n = 100) (mean ± SD; **P < 0.01, *P < 0.05; Tukey–Kramer and Bonferroni/Dunn tests)
Increase in rCTF distribution in the nuclei in a polyQ length-dependent manner
Immunoblot analysis using A6RPT-#5803 in SCA6 cerebella showed the tendency of endogenous CTF with an expanded polyQ to locate in the nucleus. To confirm if such tendency is also true in cultured cells, we assessed the frequencies of PC12 cells having nuclear rCTF expression and compared rCTF with different lengths of polyQ (Q13, Q28, or Q165). Although rCTF distribution was always predominant in the cytoplasm with any polyQs, its nuclear distribution increased in a polyQ length-dependent fashion (Q13: 0.324 ± 0.036, Q28: 0.703 ± 0.039, Q165: 0.840 ± 0.030, P < 0.001; one-way ANOVA) (Fig. 10c, d). Interestingly, the tendency for nuclear distribution was significantly strong in rCTF-Q28 compared with rCTF-Q13 (P < 0.05; Tukey–Kramer and Bonferroni/Dunn tests) (Fig. 10d), indicating that a small polyQ expansion is sufficient to cause rCTF nuclear distribution.
Discussion
One of the most important findings on immunoblot analysis is that the CTF, which is normally expressed abundantly in the cytoplasmic fraction of human cerebella, was additionally detected in SDS-insoluble/FA-soluble cytosolic fraction from SCA6 cerebella. The difference observed between control and SCA6 brains was striking considering that the difference in polyQ length was very small (5–15Q). Immunohistochemistry demonstrated abundant microscopic aggregates in SCA6 Purkinje cells but not in any control cells. We conclude from these data that a small polyQ expansion affects protein aggregation in SCA6 human brains. For recombinant proteins, rCTF-Q28 showed propensity for aggregation compared with rCTF-Q13 on immunohistochemistry in PC12 cells, whereas full-length rCav2.1 with Q28 did not. CTF oligomers were observed when rCTFs were analyzed under non-denaturing conditions. Although it is well known that polyQ expansion renders the protein containing the expansion mis-fold, our observation suggesting that the CTF behaves differently with such a small polyQ expansion both in vivo and in vitro is indeed novel and surprising. To our knowledge, this is the first study to show that a small polyQ expansion causes protein aggregation in human brains.
We detected the 75-kDa CTF in the SCA6 sucrose-soluble nuclear fraction probed with 1C2 on immunoblotting, whereas we saw relatively fewer nuclear aggregates compared to cytoplasmic ones on immunohistochemistry. This discrepancy may be brought by the differences in the protein condensations between cytoplasmic and nuclear proteins made through the present cell fractionation: the nuclear proteins were more enriched than the cytoplasmic proteins. Considering that the CTF normally expressed in the cell body as we showed in Fig. 5b, it is reasonable to consider that the nuclear CTF should be less in amount compared to the cytoplasmic CTF. In addition, aggregate formation might leave less free CTF remaining in the sucrose-soluble cytosolic fraction. Nevertheless, our data clearly showed that the CTF with expanded polyQ in a soluble form tend to mislocate in the nucleus compared to the normal CTF.
On immunoblotting of cytosolic fraction (C1), A6PRT-#5803 detected not only the 75-kDa CTF but also a band of 150 kDa, whereas only the 75-kDa CTF was detected with A6RPT-C (Fig 5b). Although the precise biochemical identity of the 150-kDa protein remains unknown, Cav2.1 proteins of similar molecular sizes have been previously demonstrated by immunoblotting with different antibodies: the antibody CT-2 detected three different bands ranging between 120 and 190 kDa in the cytosolic fraction of mice cerebella [22], and the antibody CNA3 against rat Cav2.1 detected two bands ranging between 121 and 195 kDa in rat brain protein lysates. Considering that similar proteins are detected in different species, we speculate that the 150-kDa protein could be one of the Cav2.1 isoforms generated by alternative splicing.
Since the key factor for protein aggregation is the protein context flanking the polyQ tract [3, 32] as well as the length of polyQ, it is rational to consider that the CTF contains AA sequences that may promote the CTF to aggregate. Besides its protein context, there may be other factors that cause the CTF to aggregate with such a small polyQ expansion. One of such factors is the higher expression level of Cav2.1 both in terms of mRNA and protein levels in SCA6 Purkinje cells. The CACNA1A mRNA isoforms encoding the polyQ tract are highly expressed in Purkinje cells than in any other neurons in human brains [15]. The second factor is the predirection for increase of polyQ-coding CACNA1A mRNA due to alternative splicing. It has been shown that the CAG repeat expansion enhances alternative splicing at the intron46/exon47 splice junction, resulting in much higher level of these polyQ-encoding isoforms in Purkinje cells of SCA6 cerebella [52]. Third, these could be a post-translational factor. PolyQ expansion is reported to elevate the expression levels of Cav2.1 channel protein on the plasma membrane surface of cultured cells [35]. Our data on immunoblot analysis may support these implications that Cav2.1 C-terminal region is increased in SCA6: the immunoreactive band above 250 kDa conceivably the full-length Cav2.1, and smears above 100 kDa in C2 fraction were both clearly seen in SCA6 cerebella than in control (Fig. 6a).
In several polyQ diseases, such as HD, SBMA, DRPLA, SCA2, MJD/SCA3, and SCA7, it has been well documented that production of toxic polyQ-containing fragments proteolytically cleaved from full-length proteins is apparently linked to neuronal toxicity [41, 49]. Nevertheless, it remains unclear whether cleavage of full-length proteins containing polyQ is an important step in disease pathogenesis or is a normal event in degradation of protein turnover. Concerning Cav2.1, the CTF was detected in both control and SCA6 cerebella with a comparative amount by immunoblot analysis, suggesting that the generation of CTF is based not merely on the cleavage mediated by the polyQ-expansion as in other polyQ-diseases, but on the functional nature of Cav2.1. In this context, SCA6 could be distinct from other polyQ diseases, in that the potentially toxic protein species are generated by a certain functional background. Thus, the knowledge of CTF functions would be indispensable for exploring the pathogenesis of SCA6.
Then what is the role of (Cav2.1-)CTF? The C-terminal fragments are also generated in other channel proteins, such as Cav1.2 [2], Cav1.3 [11], and Cav2.2 [58], but their roles appear distinct. The Cav1.1- and Cav1.2-CTF are known to non-covalently associate with the remainder of the channels and exert an inhibitory effect on their channel functions [12, 13]. In various neurons, the “L-type”-CTF produced from the L-type calcium channel Cav1.2 is translocated into the nucleus where it acts as a transcription factor [7]. By comparing the amino acid sequences in public database (NCBI: http://www.ncbi.nlm.nih.gov/), it was found that the Cav2.1 CTF and L-type CTF have similar molecular sizes (about 75 kDa) and AA sequences. Notably, however, the L-type CTF was shown to be expressed in the nuclei and cell bodies of rat cortical neurons [7], while the Cav2.1 CTF did not obviously localize in the nuclei under the same experimental condition (data shown upon request). This might reflect a difference in the functional properties of the Cav2.1 CTF and L-type CTF. It is known that the C-terminal region of Cav2.1 interacts with various intracellular proteins and modulates channel functions [1, 20]. Taken these facts together, it appears probable that the CTF from Cav2.1 with normal polyQ has distinct and important intracellular function(s).
To discover normal CTF functions, knowledge of its intracellular localization is a prerequisite. While Kordasiewicz et al. [22] previously showed that the CTF is mainly located in the neuronal nuclei by immunoblot analysis on mice cerebellar extracts and immunohistochemistry on control human cerebellum, we here showed that the CTF is highly expressed in the cytoplasm with the possibility of a very small amount present in the nuclei. Several explanations for this discrepancy with the previous finding of Kordasiewicz et al. may be considered. The use of different antibodies may cause such discrepancy; we used four different antibodies compared with the previously employed CT-2. The use of different cell lines is another cause; we used PC12 cells compared with the human embryonic kidney cells they used. Another compelling explanation is the use of different CTFs; we used a longer CTF consisting of C-terminal 553 AA (#1954–2506) compared with the previously utilized (515 AA, #2096–2510). If this difference was proved to be a critical factor in determining the intracellular location of the CTF, a certain sequence that is responsible for locating the CTF in the cytoplasm may be present. It would be important to note that the present rCTF had a molecular size very close to that of the CTF in human cerebella as we showed on immunoblot analysis. Based on our findings, we speculate that the CTF has a role mainly in the neuronal cytoplasm.
Another essential finding in the present study is that a small polyQ expansion not only altered CTF solubility but also affected its intracellular distribution. In contrast to control cerebella harboring normal-length polyQ ranging from Q7 to Q17, the 75-kDa CTF was clearly detected by both A6RPT-#5803 and 1C2 in the N fraction specifically from SCA6 cerebella harboring Q22. These data demonstrate that a small polyQ expansion (only 5–15Q residues longer than control polyQ) is sufficient to allow CTF detection in the nucleus. However, the precise mechanism underlying such detection remains unknown. One possible hypothesis is that the polyQ expansion may enhance CTF nuclear import by putative nuclear localization signals, as previously demonstrated in cell culture models [22]. An alternative hypothesis is that it is possible that the CTF with an expanded polyQ tends to be retained in the nucleus by forming aggregates or inhibiting nuclear export, given that the CTF is normally shuttled between the cytoplasm and the nucleus similar to the L-type CTF. These two hypotheses may not be mutually exclusive. The CTF with an expanded polyQ has been shown to cause cell death in three independent studies including ours [22, 24, 27]. One of these studies showed that such toxicity was dependent on the nuclear localization of CTF [22]. If nuclear translocation of CTF is the critical step for toxicity, interfering with the translocation could be instrumental for the fundamental therapy of SCA6.
Recently, knock-in (KI) mice that carry the CAG repeat and flanking sequences in human CACNA1A (“humanized” exon 47) were generated [57]. By immunohistochemistry with A6RPT-#5803, we demonstrated microscopic aggregates in KI mice homozygously expressing Q84 (Q84/Q84) but not in those with Q14 or Q30 [57]. Microscopic aggregates in Q84/Q84 KI mice were morphologically reminiscent of those seen in human SCA6 Purkinje cells, and located predominantly in the cell bodies of Purkinje cells. This observation not only validates the Q84/Q84 KI mice as a model for SCA6 but also supports our finding of intracellular location of aggregates. The CTF, not detected on immunoblot by CT-2 antibody [22], needs to be investigated in these mice using the present cellular fractionation technique and A6RPT-#5803 antibody. As both similarities and differences are noted in pathologies between human patients and polyQ mice model [62], further investigations on SCA6KI mice with careful comparisons with human patients would be an important approach.
Based on the present observations in human brains, we highlight some unique neuropathological features of SCA6 that could be important for dissecting the pathogenesis of this disease. First, the Purkinje cell has been regarded as a neuron particularly devoid of aggregate formation in other polyQ diseases. For instance, Purkinje cells showed neuronal loss without any inclusions in SCA1, SCA2, MJD/SCA3, and DRPLA [23]. On the other hand, we showed here that Cav2.1 built aggregate both selectively and abundantly in this neuron. Therefore, a clear correlation is seen in SCA6 between distribution of mutant protein aggregates and degeneration, whereas this match is not complete, or even paradoxical, in other polyQ diseases. This might indicate that the aggregate formation is particularly important for the SCA6 pathogenesis. Second, our observation suggested that the CTF is one of the important protein species forming aggregation. Considering that the CTF is also present in controls with still unidentified, but potentially crucial, cellular functions [4], it is possible that the SCA6 pathogenesis is deeply associated with the CTF functions. For example, it is possible that cytoplasmic aggregations of CTF may lead to cellular dysfunction via reduction of proper CTF to play such essential functions and thus compromising normal functions of CTF. Establishing the CTF knock-down animal model is necessary to clarify this possibility. Precise knowledge of the C-terminal region of Cav2.1 is essential for understanding SCA6. It is also possible that the CTF may gain a new function by having an expanded polyQ (gain-of-function mechanism). Increased binding to myosin2B with expansion of polyQ [27] is an example for such hypothesis. Other interacting proteins, such as II–III-loop of Cav2.1, G-proteins, may also play some roles in pathogenesis [56]. It is also conceivable that formation of abundant aggregates in cell bodies may disturb normal cellular functions such as intracellular trafficking [9]. Finally, an increase in nuclear CTF localization may affect normal nuclear function, considering the previous work expressing the CTF resulted in cell death [22]. Although SCA6 do not show prominent intranuclear aggregates or diffuse nuclear staining described in other polyQ diseases [63], it is possible to consider that SCA6 conforms to other polyQ diseases in that soluble polyQ-containing protein exists in the nucleus. A line of evidence suggests that nuclear inclusions in other polyQ diseases may even act to protect neurons from polyQ-mediated toxicity [54], while a soluble monomer or oligomers of expanded polyQ-containing proteins are more toxic to cells [31, 46]. Thus, it would be important whether the CTF may lead to nuclear dysfunction. Because the polyQ length is small in SCA6, the nuclear dysfunction such as CREB-dependent transcription alteration may not be working in SCA6 [29, 42]. These several major speculations may not be mutually exclusive, suggesting that pathogenic mechanisms of SCA6 could be complicated. Additional studies are needed to further clarify the functional roles of a normal CTF and to discover how the CTF with an expanded polyQ causes neurodegeneration.
References
Catterall WA (2000) Structure and regulation of voltage-gated Ca2+ channels. Annu Rev Cell Dev Biol 16:521–555
De Jongh KS, Colvin AA, Wang KK, Catterall WA (1994) Differential proteolysis of the full-length form of the L-type calcium channel alpha 1 subunit by calpain. J Neurochem 63:1558–1564
Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S (2006) Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci USA 103:11045–11050
Fletcher CF, Lutz CM, O’Sullivan TN et al (1996) Absence epilepsy in tottering mutant mice is associated with calcium channel defects. Cell 87:607–617
Frontali M (2001) Spinocerebellar ataxia type 6: channelopathy or glutamine repeat disorder? Brain Res Bull 56:227–231
Gierga K, Schelhaas HJ, Brunt ER et al (2009) Spinocerebellar ataxia type 6 (SCA6): neurodegeneration goes beyond the known brain predilection sites. Neuropathol Appl Neurobiol 35:515–527
Gomez-Ospina N, Tsuruta F, Barreto-Chang O, Hu L, Dolmetsch R (2006) The C terminus of the L-type voltage-gated calcium channel Ca(V)1.2 encodes a transcription factor. Cell 127:591–606
Gomez CM, Thompson RM, Gammack JT et al (1997) Spinocerebellar ataxia type 6: gaze-evoked and vertical nystagmus, Purkinje cell degeneration, and variable age of onset. Ann Neurol 42:933–950
Gunawardena S, Goldstein LS (2005) Polyglutamine diseases and transport problems: deadly traffic jams on neuronal highways. Arch Neurol 62:46–51
Hazeki N, Tukamoto T, Goto J, Kanazawa I (2000) Formic acid dissolves aggregates of an N-terminal huntingtin fragment containing an expanded polyglutamine tract: applying to quantification of protein components of the aggregates. Biochem Biophys Res Commun 277:386–393
Hell JW, Westenbroek RE, Warner C et al (1993) Identification and differential subcellular localization of the neuronal class C and class D L-type calcium channel alpha 1 subunits. J Cell Biol 123:949–962
Hulme JT, Konoki K, Lin TW et al (2005) Sites of proteolytic processing and noncovalent association of the distal C-terminal domain of CaV1.1 channels in skeletal muscle. Proc Natl Acad Sci USA 102:5274–5279
Hulme JT, Yarov-Yarovoy V, Lin TW, Scheuer T, Catterall WA (2006) Autoinhibitory control of the CaV1.2 channel by its proteolytically processed distal C-terminal domain. J Physiol 576:87–102
Ikeuchi T, Takano H, Koide R et al (1997) Spinocerebellar ataxia type 6: CAG repeat expansion in alpha1A voltage-dependent calcium channel gene and clinical variations in Japanese population. Ann Neurol 42:879–884
Ishikawa K, Fujigasaki H, Saegusa H et al (1999) Abundant expression and cytoplasmic aggregations of [alpha]1A voltage-dependent calcium channel protein associated with neurodegeneration in spinocerebellar ataxia type 6. Hum Mol Genet 8:1185–1193
Ishikawa K, Owada K, Ishida K et al (2001) Cytoplasmic and nuclear polyglutamine aggregates in SCA6 Purkinje cells. Neurology 56:1753–1756
Ishikawa K, Tanaka H, Saito M et al (1997) Japanese families with autosomal dominant pure cerebellar ataxia map to chromosome 19p13.1–p13.2 and are strongly associated with mild CAG expansions in the spinocerebellar ataxia type 6 gene in chromosome 19p13.1. Am J Hum Genet 61:336–346
Ishikawa K, Watanabe M, Yoshizawa K et al (1999) Clinical, neuropathological, and molecular study in two families with spinocerebellar ataxia type 6 (SCA6). J Neurol Neurosurg Psychiatry 67:86–89
Jodice C, Mantuano E, Veneziano L et al (1997) Episodic ataxia type 2 (EA2) and spinocerebellar ataxia type 6 (SCA6) due to CAG repeat expansion in the CACNA1A gene on chromosome 19p. Hum Mol Genet 6:1973–1978
Kitano J, Nishida M, Itsukaichi Y et al (2003) Direct interaction and functional coupling between metabotropic glutamate receptor subtype 1 and voltage-sensitive Cav2.1 Ca2+ channel. J Biol Chem 278:25101–25108
Kordasiewicz HB, Gomez CM (2007) Molecular pathogenesis of spinocerebellar ataxia type 6. Neurotherapeutics 4:285–294
Kordasiewicz HB, Thompson RM, Clark HB, Gomez CM (2006) C-termini of P/Q-type Ca2+ channel alpha1A subunits translocate to nuclei and promote polyglutamine-mediated toxicity. Hum Mol Genet 15:1587–1599
Koyano S, Iwabuchi K, Yagishita S, Kuroiwa Y, Uchihara T (2002) Paradoxical absence of nuclear inclusion in cerebellar Purkinje cells of hereditary ataxias linked to CAG expansion. J Neurol Neurosurg Psychiatry 73:450–452
Kubodera T, Yokota T, Ohwada K et al (2003) Proteolytic cleavage and cellular toxicity of the human alpha1A calcium channel in spinocerebellar ataxia type 6. Neurosci Lett 341:74–78
Kuroiwa K, Li H, Tago K et al (2000) Construction of ELISA system to quantify human ST2 protein in sera of patients. Hybridoma 19:151–159
Lunkes A, Lindenberg KS, Ben-Haiem L et al (2002) Proteases acting on mutant huntingtin generate cleaved products that differentially build up cytoplasmic and nuclear inclusions. Mol Cell 10:259–269
Marqueze-Pouey B, Martin-Moutot N, Sakkou-Norton M et al (2008) Toxicity and endocytosis of spinocerebellar ataxia type 6 polyglutamine domains: role of myosin IIb. Traffic 9:1088–1100
Matsuyama Z, Kawakami H, Maruyama H et al (1997) Molecular features of the CAG repeats of spinocerebellar ataxia 6 (SCA6). Hum Mol Genet 6:1283–1287
McCampbell A, Taylor JP, Taye AA et al (2000) CREB-binding protein sequestration by expanded polyglutamine. Hum Mol Genet 9:2197–2202
Mori Y, Friedrich T, Kim MS et al (1991) Primary structure and functional expression from complementary DNA of a brain calcium channel. Nature 350:398–402
Nagai Y, Inui T, Popiel HA et al (2007) A toxic monomeric conformer of the polyglutamine protein. Nat Struct Mol Biol 14:332–340
Nozaki K, Onodera O, Takano H, Tsuji S (2001) Amino acid sequences flanking polyglutamine stretches influence their potential for aggregate formation. Neuroreport 12:3357–3364
Ophoff RA, Terwindt GM, Vergouwe MN et al (1996) Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutations in the Ca2+ channel gene CACNL1A4. Cell 87:543–552
Peters MF, Ross CA (1999) Preparation of human cDNas encoding expanded polyglutamine repeats. Neurosci Lett 275:129–132
Piedras-Renteria ES, Watase K, Harata N et al (2001) Increased expression of alpha 1A Ca2+ channel currents arising from expanded trinucleotide repeats in spinocerebellar ataxia type 6. J Neurosci 21:9185–9193
Restituito S, Thompson RM, Eliet J et al (2000) The polyglutamine expansion in spinocerebellar ataxia type 6 causes a beta subunit-specific enhanced activation of P/Q-type calcium channels in Xenopus oocytes. J Neurosci 20:6394–6403
Saegusa H, Wakamori M, Matsuda Y et al (2007) Properties of human Cav2.1 channel with a spinocerebellar ataxia type 6 mutation expressed in Purkinje cells. Mol Cell Neurosci 34:261–270
Sakurai T, Westenbroek RE, Rettig J, Hell J, Catterall WA (1996) Biochemical properties and subcellular distribution of the BI and rbA isoforms of alpha 1A subunits of brain calcium channels. J Cell Biol 134:511–528
Sasaki H, Kojima H, Yabe I et al (1998) Neuropathological and molecular studies of spinocerebellar ataxia type 6 (SCA6). Acta Neuropathol 95:199–204
Scherzinger E, Lurz R, Turmaine M et al (1997) Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 90:549–558
Shao J, Diamond MI (2007) Polyglutamine diseases: emerging concepts in pathogenesis and therapy. Hum Mol Genet 16(special issue 2):R115–R123
Shimohata T, Nakajima T, Yamada M et al (2000) Expanded polyglutamine stretches interact with TAFII130, interfering with CREB-dependent transcription. Nat Genet 26:29–36
Stea A, Tomlinson WJ, Soong TW et al (1994) Localization and functional properties of a rat brain alpha 1A calcium channel reflect similarities to neuronal Q- and P-type channels. Proc Natl Acad Sci USA 91:10576–10580
Takahashi H, Ikeuchi T, Honma Y, Hayashi S, Tsuji S (1998) Autosomal dominant cerebellar ataxia (SCA6): clinical, genetic and neuropathological study in a family. Acta Neuropathol 95:333–337
Takahashi H, Ishikawa K, Tsutsumi T et al (2004) A clinical and genetic study in a large cohort of patients with spinocerebellar ataxia type 6. J Hum Genet 49:256–264
Takahashi T, Kikuchi S, Katada S et al (2008) Soluble polyglutamine oligomers formed prior to inclusion body formation are cytotoxic. Hum Mol Genet 17:345–356
Takano H, Cancel G, Ikeuchi T et al (1998) Close associations between prevalences of dominantly inherited spinocerebellar ataxias with CAG-repeat expansions and frequencies of large normal CAG alleles in Japanese and Caucasian populations. Am J Hum Genet 63:1060–1066
Tanaka O, Sakagami H, Kondo H (1995) Localization of mRNAs of voltage-dependent Ca(2+)-channels: four subtypes of alpha 1- and beta-subunits in developing and mature rat brain. Brain Res Mol Brain Res 30:1–16
Tarlac V, Storey E (2003) Role of proteolysis in polyglutamine disorders. J Neurosci Res 74:406–416
Toru S, Murakoshi T, Ishikawa K et al (2000) Spinocerebellar ataxia type 6 mutation alters P-type calcium channel function. J Biol Chem 275:10893–10898
Trottier Y, Lutz Y, Stevanin G et al (1995) Polyglutamine expansion as a pathological epitope in Huntington’s disease and four dominant cerebellar ataxias. Nature 378:403–406
Tsunemi T, Ishikawa K, Jin H, Mizusawa H (2008) Cell-type-specific alternative splicing in spinocerebellar ataxia type 6. Neurosci Lett 447:78–81
Tsunemi T, Saegusa H, Ishikawa K et al (2002) Novel Cav2.1 splice variants isolated from Purkinje cells do not generate P-type Ca2+ current. J Biol Chem 277:7214–7221
Uchihara T, Iwabuchi K, Funata N, Yagishita S (2002) Attenuated nuclear shrinkage in neurons with nuclear aggregates–a morphometric study on pontine neurons of Machado-Joseph disease brains. Exp Neurol 178:124–128
Vacher H, Mohapatra DP, Trimmer JS (2008) Localization and targeting of voltage-dependent ion channels in mammalian central neurons. Physiol Rev 88:1407–1447
Walker D, De Waard M (1998) Subunit interaction sites in voltage-dependent Ca2+ channels: role in channel function. Trends Neurosci 21:148–154
Watase K, Barrett CF, Miyazaki T et al (2008) Spinocerebellar ataxia type 6 knockin mice develop a progressive neuronal dysfunction with age-dependent accumulation of mutant CaV2.1 channels. Proc Natl Acad Sci USA 105:11987–11992
Westenbroek RE, Hell JW, Warner C et al (1992) Biochemical properties and subcellular distribution of an N-type calcium channel alpha 1 subunit. Neuron 9:1099–1115
Westenbroek RE, Sakurai T, Elliott EM et al (1995) Immunochemical identification and subcellular distribution of the alpha 1A subunits of brain calcium channels. J Neurosci 15:6403–6418
Williams AJ, Paulson HL (2008) Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci 31:521–528
Yabe I, Sasaki H, Matsuura T et al (1998) SCA6 mutation analysis in a large cohort of the Japanese patients with late-onset pure cerebellar ataxia. J Neurol Sci 156:89–95
Yamada M, Sato T, Tsuji S, Takahashi H (2008) CAG repeat disorder models and human neuropathology: similarities and differences. Acta Neuropathol 115:71–86
Yamada M, Tsuji S, Takahashi H (2000) Pathology of CAG repeat diseases. Neuropathology 20:319–325
Yue Q, Jen JC, Nelson SF, Baloh RW (1997) Progressive ataxia due to a missense mutation in a calcium-channel gene. Am J Hum Genet 61:1078–1087
Zhuchenko O, Bailey J, Bonnen P et al (1997) Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the alpha 1A-voltage-dependent calcium channel. Nat Genet 15:62–69
Acknowledgments
We thank Dr. Kei Watase (Tokyo Medical and Dental University) for his stimulating discussions and generous help, and Ms. Iku Sudo, Minori Kono and Hideko Uno for their technical assistance. We acknowledge Drs. Toshiki Uchihara (Tokyo Metropolitan Institute for Neuroscience, Fuchu, Japan) and Yuko Saito (National Center of Neurology and Psychiatry, Kodaira, Japan) for critically reading the manuscript, and Scientific Language Editing Team, Tsukuba, Japan for professionally editing the manuscript. This study was funded by the Japanese Ministry of Education, Sports and Culture (K.I. and H.M.), the Japan Society for Promotion of Science (JSPS) (K.I. and H.M.), the 21st Century COE Program on Brain Integration and its Disorders from the Japanese Ministry of Education, Science and Culture (H.M.), and from the Health and Labour Sciences Research Grants on Human Genome (K.I.) and Ataxic Diseases (H.M.) of the Japanese Ministry of Health, Labour and Welfare.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Ishiguro, T., Ishikawa, K., Takahashi, M. et al. The carboxy-terminal fragment of α1A calcium channel preferentially aggregates in the cytoplasm of human spinocerebellar ataxia type 6 Purkinje cells. Acta Neuropathol 119, 447–464 (2010). https://doi.org/10.1007/s00401-009-0630-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-009-0630-0