Abstract
Sorgoleone is a secondary sorghum metabolite released from roots. It has allelopathic properties and is considered to inhibit ammonia-oxidizing archaea (AOA) and bacteria (AOB) responsible for the rate-limiting step (ammonia oxidation) in nitrification. Low activity of these microorganisms in soil may contribute to slow down nitrification and reduce nitrogen loss via denitrification and NO3− leaching. The potential nitrification rate (PNR) and the composition of microbial communities were monitored in rhizosphere soil to investigate the growth effect sorghum on biological nitrification inhibition (BNI). A greenhouse pipe experiment was conducted using sorghum lines IS20205 (high-sorgoleone release ability), IS32234 (medium-sorgoleone release ability), 296B (low-sorgoleone release ability), and a control (no plants) combined with fertilization application of 0 or 120 kg N ha−1. We applied nitrogen as ammonium sulfate at 16 days (20 N), 37 days (40 N), and 54 days (60 N). We collected soil solutions at 7.5 cm depths every 3 days and measured the pH and nitrate levels. At 1 and 2.3 months, we sampled the bulk and rhizosphere soils and roots in the 0–10 cm, 10–30 cm, and 30–80 cm depths to determine NO2, mineral N, total N, total C, sorgoleone, the composition of AOA, AOB, and total bacteria and archaea. Sorgoleone was continuously released throughout the 2.3 months’ growth and was significantly higher in IS20205, followed by IS32234 then 296B, which showed shallow levels. The IS2020 5rhizosphere showed lower NO2 and nitrate levels and significant inhibition of AOA populations. However, we did not find significant differences in the abundance of AOB between plant treatments. Multivariate analysis and Spearman’s correlations revealed that sorgoleone as well as environmental factors such as soil pH, soil moisture, NO3−-N, and NH4+-N shape the composition of microbial communities. This study demonstrated that the release of higher amounts of sorgoleone has great potential to inhibit the abundance of AOA and soil nitrification. The breeding of sorghum lines with the ability to release higher amounts of sorgoleone could be a strategic way to improve the biological nitrification inhibition during cultivation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nitrification is the key and often the rate-limiting step in the N cycle, which includes two steps, i.e., oxidation of ammonia (NH4+) to nitrite (NO2−) mediated by ammonia-oxidizing bacteria (AOB) (Purkhold et al. 2000; Treusch et al. 2005) and archaea (AOA) (Könneke et al. 2005) and nitrite to nitrate (NO3−) by nitrite-oxidizing bacteria (NOB) mainly from the genus Nitrobacter (Hayatsu et al. 2008). The ammonia monooxygenase (AMO) enzyme, one of the key enzymes responsible for the conversion of ammonia to nitrite, is encoded by the ammonia monooxygenase genes amoA, amoB, and amoC (Klotz et al. 1997). Among these genes, the amoA gene had been more extensively used for the study of ammonia oxidizers and has a well-established database, compared with amoB and amoC (Francis et al. 2005; Okano et al. 2004).
Understanding the community composition and spatial distribution of AOA and AOB is essential for assessing the nitrification process (Li et al. 2018) since these two groups are responsible for the first-step (ammonia oxidation) in nitrification. Therefore, finding ways to maintain the abundance and more specifically the activity of amoA genes at low levels is vital to slow down the nitrification and reduce N loss via denitrification (emissions of N2O and NO), nitrate leaching, and improve N use efficiency (NUE). Nitrate leaching in agricultural soils is estimated to be as high as 61.5 Tg N year−1 (Schlesinger 2009). The NUE for global cereal production is estimated at 33% (Raun and Johnson 1999). The control of the nitrification process in soil is necessary to minimize N losses and increase the NUE in agricultural fields (Subbarao et al. 2015). Synthetic nitrification inhibitors such as thiourea and dicyandiamide (DCD) have been developed (Prasad and Power 1995), but their widespread use in agriculture is limited due to high costs and variable performance across different agro-climatic and soil environments (Nardi et al. 2013). Subbarao et al. (2009) reported that under these ecosystems, nitrification could potentially lead to N starvation for plants, which in turn forces them to develop strategies to protect N from loss. Some plants have developed mechanisms to suppress soil nitrifier activity, a plant function termed “biological nitrification inhibition (BNI)” (Subbarao et al. 2006a). To date, several BNI plants have been identified, including Brachiaria pasture grasses (Ishikawa et al. 2003; Subbarao et al. 2007; Jones 2013) and sorghum (Subbarao et al. 2013). A bioluminescence assay using a recombinant Nitrosomonas europaea strain (harboring luxAB genes from Vibrio harveyi) has been developed and extensively used to estimate the BNI potential of root exudates of several crops including sorghum (Iizumi et al. 1998; Subbarao et al. 2006b; Zakir et al. 2008).
Sorghum is the fifth most widely cultivated cereal grain globally, grown mainly in drought-prone and less developed regions of the arid and semi-arid tropics (Smith and Frederiksen 2000; Subudhi and Nguyen 2000). This crop is well known to be producing a range of metabolites having allelopathic or phytotoxic effects, and in some cases, the phytotoxic effect could be attributed to specific compounds like sorgoleone (Einhellig and Souza 1992). Sorgoleone is an oily brownish exudate that contains lipid benzoquinones, which constitutes > 90% of the hydrophobic components of sorghum root exudates (Netzly and Butler 1986; Czarnota et al. 2003). It has been shown to inhibit nitrifier activity (Subbarao et al. 2013), and together with the phenolic substance methyl 3-(4-hydroxyphenyl) propionate (MHPP) (Zakir et al. 2008) and sakuranetin (Subbarao et al. 2013), they form the major BNIs released from sorghum roots. However, most studies addressing the effect of sorgoleone on nitrifiers and soil nitrification have been carried out under laboratory incubation at short-term scales. Very few have closely investigated the dynamic of sorgoleone release throughout the plant’s growth cycle in natural conditions and how its release timely affects AOA and AOB communities and soil nitrification in the presence or absence of N fertilization.
In this study, three sorghum genetic stocks and a control (no plant) were grown without and with N fertilization (120 N) for 2.3 months. The aim was to determine (1) how the growth of sorghum genetic stocks that release sorgoleone at different levels affects the composition of not only ammonia-oxidizing microbes (AOB, AOA) but also that of abundances of bacteria and archaea?; (2) whether both AOA and AOB abundances affected by the release of sorgoleone in natural conditions and at which magnitude?; and (3) how the interaction between sorgoleone release and soil nitrifiers shapes the soil potential nitrification and how this affects plant growth? The study was performed using a “pipe experiment” setup in a greenhouse. Microbial composition and abundance and were analyzed using quantitative real-time polymerase chain reaction (qPCR) and Illumina MiSeq targetting the amoA genes for AOB and AOA and V4 region of 16S rRNA genes for total bacteria and archaea. Soil chemical analysis complemented these molecular analyses, and we interpreted all data in a holistic approach.
Materials and methods
We set up the experiment in a greenhouse (Tm: 29.6 °C average, RH: 63.8%) using ϕ 12 cm × 1 m pipes (Hokkaido Ltd., Japan) filled with soil, herein referred to as “pipe experiment.”
Soil preparation
The soil used was Andosol collected from the Japan International Research Center for Agricultural Sciences (JIRCAS) experimental station (Hachimandai) in Tsukuba, Japan (36.0535° N and 140.0792° E). The soil was not cultivated over many years nor received any fertilization before its collection. Before use, it was well mixed and passed through a 5-mm sieve to remove big-size soil particles. Original water content (47%), percent moisture at field capacity (61%), and other soil properties were measured immediately after collection, and 12.5 kg fresh soil (8.5 kg oven-dry weight) was added in each pipe. Before, we calculated the amount of water in a pipe when at field capacity (FC) and the required water volume to add during each watering time to adjust the moisture level to that under FC.
Treatments and experimental design
The treatments consisted of two factors, F1 and F2. F1 corresponded to 3 sorghum lines with contrasting sorgoleone release ability (IS20205 19.11 μg/mg dry root, IS32234 19.12 μg/mg dry root, 296B 5.16 μg/mg dry root; Subbarao GV, JIRCAS, unpublished results) and a control treatment (no plant). Sorgoleone was initially determined on 7-day-old sorghum seedlings as described later in this paper. We recieved the sorghum seeds from ICRISAT (Pantancheru, India). The second factor F2 corresponded to a fertilization regime without and with ammonium sulfate (120 kg N ha−1). The resulting eight treatments (F1 × F2) were replicated eight times and arranged in a completely randomized block design. In total, there were four blocks containing 16 pipes (F1 (4) × F2 (2) × 2 harvesting periods) each. However, an extra pipe was added per block to assess the amount of water to add during watering.
Before adding the soil, a 0.15-mm-thick cylindrical plastic, with a similar diameter size than the pipes, was sealed at the bottom and inserted inside each pipe. After adding the soil inside the plastics inserted in the pipes, pipes were hit on the soil surface up and down ten times with a similar force to create an identical bulk density and soil distribution. At 7.5 cm horizon from the soil surface of each pipe, a 10-cm porous ceramic rhizon cup (Rhizosphere Research Products, Wageningen, the Netherlands) was fully buried horizontally in the soil of each pipe. At the contact zone with the pipe, we inserted the rhizon in a suction tube (10 cm length, 4 mm Ø) connected to a 10-ml vacuum syringe (Fig. S1). The soil solutions were sucked every 3 days and collected in the syringes, 1 h after watering the pipe to field capacity, and the extracts were used to determine the nitrates (NO3−) levels and pH variation at the topsoil of the different treatments.
Fertilization and watering
Both K and P were applied as basal fertilization to each pipe at 150 kg K2O ha−1 (124.5 kg K ha−1) and 150 kg P2O5 ha−1 (65.55 kg P ha−1), based on pipe surface area (PSA = π × r2) corresponding to 122.72 cm2. The amount of each fertilizer required per pipe (122.72 cm2) was calculated and dissolved in 1.2 l (volume of water to adjust to FC) and then applied to the pipe. On the following day, six surface-sterilized (soaking in 5% sodium hypochlorite for 10 min, 99.9% ethanol for 3 min, followed by five washes with sterilized deionized water) sorghum seeds per line were sown in corresponding pipes. After sowing, we recorded the total weight of the four extra pipes (3 lines, one control) called after planting target weight (APTW), and this weight was aimed at each watering time. At later growth stages, an approximate extra weight representing the plant biomass was added to the APTW when estimating the amount of water for watering. Three homogenous seedlings per pipe were left 1 week after sowing. Ammonium sulfate (120 kg N ha−1) was applied to the corresponding pipes at 16 days after sowing (DAS) at the rate of 20 kg N ha−1 (20 N), 37 DAS (40 N), and 54 DAS (60 N), after dissolving the required amounts in a solution equalling the decreased amount of water at the watering day. Deionized water was used for watering to avoid contamination with other nutrients.
Plant harvesting and soil sampling
Four replicates per treatments were destructively sampled at 31 and 32 DAS (1 month), and the remaining pipes were harvested at 70 and 71 DAS (2.3 months). Shoots were collected and oven-dried at 75 °C for 48 h to record the dry weights. Before soil and root sampling, the rhizon tubes were first carefully removed, and the plastic containing the soil and root column was pulled off the pipe. We used a clean cutter to separate the three soil cores (0–10 cm, 10–30 cm, 30–80 cm horizons) per pipe. All the soil in control’s cores is referred to as bulk soil. Cores containing plant roots were shaken by hand to release the loose soil, and the rhizosphere soil corresponding to the soil finely attached to roots after releasing the loose soil was collected on clean trays using clean brushes. Sub-samples from bulk soils (about 40 g) and rhizosphere soils per core were put in 50-ml falcon tubes and transferred to the laboratory in cold conditions (4–10 °C) and used for chemical and molecular analyses. After removing the rhizosphere soil, the weight of fresh roots per core was immediately recorded. Each core’s root biomass was put in plastic bags and transferred to the laboratory in the same conditions as above. On the same day, sorgoleone was extracted from roots as described by Tesfamariam et al. (2014) with the only difference that roots were dipped into 100 mL acidified dichloromethane or DCM (1% acetic acid:DCM v/v) instead of 20 mL due to their higher amounts. The filtrates were stored at 4 °C until sorgoleone quantification. After the DCM wash, the roots were again washed with tap water and cleaned with deionized water to remove the remaining soil particles. They were oven-dried at 70 °C for 72 h to calculate the total root dry weight per soil core and per plant.
Quantification of sorgoleone release ability from sorghum roots at harvests (1 and 2.3 months)
Briefly, the filtered acidified root-DCM washes stored 4 °C were evaporated to dryness using a rotary evaporator at 40 °C, 95 rpm. The residues were treated with 20 mL methanol and evaporated again. The new residues were harvested with another 1.5 mL methanol into a 2-mL Eppendorf tube and then filtered through a 0.20-μm syringe membrane filter to remove any insoluble compounds that might interfere with HPLC analysis. We transferred 150 μl filtrate into a 0.6-ml tube and evaporated it to dryness after making a hole in the lid of the tube. After evaporation, the residues were dissolved in 150 μl acetonitrile for sorgoleone (μg g−1 dry root) quantification by HPLC (Subbarao et al. 2013; Tesfamariam et al. 2014). The remaining 1.35 ml methanol filtrates were stored at 4 °C for further uses.
Chemical analysis
NO3− in water extracts
A standard curve was first made by plotting the absorbance reads of a series of 11 dilutions using 100 mg NO3-N l−1 standard (Wako Pure Chemical Industries, Ltd., Japan). An aliquot (1.3 ml) of the 100 mg NO3-N l−1 standard and the diluted solutions including a blank were added in a NO3 pack test tube (Kyoritsu Chemical-Check Lab., Crop., Japan) and vortexed for 10 s. The absorbance of the purple color developed after 10 min incubation was read spectrophotometrically at 540 nm wavelength. The standard curve with an equation correlating the obtained absorbance values and the corresponding NO3-N concentrations was made. Similarly, 1.3 ml water extracts were added in a pack test tube, vortexed for 10 s, and the absorbance read at 540 nm. The equation obtained in the standard curve was then used to estimate the NO3-N amounts in soil extracts. The NO3-N concentrations of extracts were assessed at each sampling time. The pH of water extracts was directly read on a pH meter.
NO3−, NH4+, potential nitrification (NO2−), pH, TC, and TN in soils
Three fresh soil replicates (1 g) of known moisture content (initial soil, bulk, and rhizosphere soils) were placed in a 50-ml falcon tube, and 10 ml 1 M KCl was added. Tubes were incubated at 150 rpm, at 25 °C, for 30 min, and 4 ml per sample was centrifuged at 15,000×g, at 25 °C, for 5 min. NO3-N concentration (μg NO3-N g−1 soil) was determined as above, taking into consideration the %moisture of the used soil and the isotopic equivalence of N. Using 625 μl of the 1 M KCl extract, NH4+ concentration (μg NH4-N g−1 soil) was determined colorimetrically according to the sodium salicylate method (Forster 1995).
The potential nitrification rate (PNR) was determined on the same day of soil sampling. Briefly, 0.5 g of fresh soils was added to 50-ml falcon tubes, and 12 ml of phosphate buffer solutions (1 mM KH2PO4, 1 mM (NH4)2SO4, 10 mM NaClO3, and pH adjusted to 7.0) was added. The tubes were shaken by hand for about 5 s, and 2 ml solution was centrifuged at 15,000×g, 25 °C, for 5 min (0 h). The remaining solution was incubated for 4 h at 25 °C, 180 rpm, after which another 2 ml solution was centrifuged as above. NO2-N was determined following the same method as NO3-N with the difference that NO2 pack test tubes were used instead of NO3 pack test tubes. A standard curve as in NO3− was also made using a 1000-mg l−1 NO2-N standard (Wako Pure Chemical Industries, Ltd., Japan). The potential nitrification (nmol NO2-N g−1 soil h−1) was calculated following Morimoto et al. (2011), taking into account the absorbance values obtained at 0 and 4 h incubation, the isotopic equivalence of N (14), and the percent moisture of the soil. Since Belser and Mays (1980) showed that the nitrite oxidized was linear from 0 h to 1 day, we set up our incubation time from 0 to 4 h, while Morimoto et al. (2011) incubated for just 2 h. Finally, soil pH was measured using a 1:2 ratio of soil to H2O. Soil samples in the 0–10 cm and 10–30 cm were selected for the determination of total N and total C by the dry combustion method using an NC analyzer (Sumigraph NC-220: Sumika Chemical Analysis Service, Osaka, Japan).
Molecular analysis
In this study, we followed the guidelines of two opinion papers about sampling strategies, DNA extraction, metadata collection (Vestergaard et al. 2017), processing, and interpretation of amplicon sequence data (Schöler et al. 2017).
DNA extraction and qPCR of bacterial and archaeal 16S rRNA, AOB, and AOA amoA genes
Soil DNA was extracted from 0.4 g fresh soil using Fast DNA Spin Kit for Soil (Mo-Bio, Carlsbad, CA) according to the manufacturer’s instructions, with a slight modification. Briefly, for better Andosol’s DNA recovery, we added 160 μl of 20% skim milk to 818 μl sodium phosphate buffer (SPB) to have the 978 μl recommended SPB. The addition of skim milk has been suggested to improve the DNA extraction from soils that strongly adsorb DNA such as Andosols (Takada-Hoshino and Matsumoto 2004, 2005). The obtained pure DNA was quantified by Qubit Quantification Platform dsDNA HS Assay kit (Invitrogen, Carlsbad, CA). Quantitative PCR (qPCR) was performed to assess the abundance of the amoA genes of both ammonia-oxidizing bacteria (AOB) and ammonia-oxidizing archaea (AOA), as well as the bacterial and archaeal 16S rRNA gene. The qPCR was performed in 15-μl reaction mixtures containing the following components: 7.5 μl of SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, BioRad Laboratories, Inc., Hercules, CA), 0.25 μM of each primer (corresponding to 0.0375 μl of 100 pmol μl−1), 1 μl of one-tenth diluted DNA, and 6.425 μl sterilized Milli-Q water. Reactions were run in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad) using the primers and the thermo-cycling conditions reported in Table 1a. Before qPCR of extracted DNA samples, replicated negative controls (DNA replaced by water) were run to verify the purity of the used kits and solutions. Since no amplification was observed, the DNA samples were solely run in the subsequent reactions. Plasmid standards of these different genes were made after PCR amplification of the extracted DNA with the same primers (Table 1a). Purified amplicons were cloned into the pGEM T-easy vector system I (Promega, Madison, WI, USA) and transformed into competent cells of Escherichia coli JM109. Plasmids of positive clones were extracted and quantified on a Nanodrop 2000 (Thermo Scientific, USA), and the copy numbers of the different genes were calculated with the concentration of the extracted plasmid DNA. The standard curves were generated using 10-fold serial dilutions of a plasmid containing the target gene insert. First, gene copy numbers were generated using a regression equation that converts the cycle threshold (Ct) value to the known number of copies in the standards. Following that, copy numbers were converted into per gram of dry soil by the following formula: \( \mathrm{Copy}\ \mathrm{number}=\frac{(Xd)\times V}{0.4\times \left(1-M\right)} \), where X indicates the copy numbers of genes detected by qPCR, d indicates the dilution factor, V indicates the volume (μl) of extracted DNA, 0.4 indicates the amount of soil used for DNA extraction (g), and M indicates soil moisture. For all assays, amplification efficiency ranged between 90 and 100% and r2 values was 0.993–0.999. DNA was extracted in duplicate per soil samples, and qPCR reactions were run in duplicate for each DNA sample.
Illumina MiSeq
Extracted soil DNA from the 0–10 cm samples was selected for sequencing by Illumina MiSeq. For total bacteria and archaea, 24 DNA samples (8 treatments × 3 replicates) from the first harvest (1 month) and 24 samples from the second harvest (2.3 months) were analyzed, targetting the V4 hypervariable region of the 16S RNA gene using the 515F-modified (Parada et al. 2016)/806R-modified (Apprill et al. 2015) primer pair. For the AOB amoA gene, we used 24 samples from the first harvest, and for AOA amoA, the 24 samples were selected from the second harvest. The primer sets for Illumina MiSeq of AOB amoA and AOA amoA genes were the same as those used during qPCR. Each gene was first amplified in a 10-μl reaction (Sarr et al. 2019) using in a thermal cycler (iCycler BioRad, Hercules, CA), following the PCR conditions described in Table 1b. The purified amplicons were used as templates (10 ng) for the second PCR, using the primer pair 2ndF and 2ndR and same PCR conditions as described in Sarr et al. (2019). These second primers include each an 8-bp barcode sequence (Index2 and Index1), allowing for multiplexing multiple samples. Amplicons were purified by AMPure XP (BECKMAN COULTER), and Synergy H1 and QuantiFluor dsDNA System estimated final library concentrations. Libraries were sequenced on paired-end 2 × 300 bp Illumina MiSeq runs at the Bioengineering Lab, Co., Ltd. (Kanagawa, Japan).
Bioinformatics
Using the Fastx toolkit of fastq_barcode_spliltter (Hannon 2010), only reads whose sequences matched precisely with the used primers were extracted. Primer sequences were deleted for all reactions, and for the V4 region of 16S rRNA, the last 50 bases of each read were also deleted. Then, using sickle tools, sequences with a quality value of less than 20 were removed, and sequences with a length of 40 bases or less and their paired sequences were discarded. Using the pair-end merge script FLASH (Magoc and Salzberg 2011), the quality-filtered sequences were merged. For V4, a fragment length of 260 bp was obtained after merging the reads (230 bp each) and removing the overlapping region with a minimum length of 10 bases. The filtered reads of V4 were checked for chimera sequences using the usearch_61 algorithm (Edgar 2010), and all sequences that were not judged to be chimeras were extracted and used for further analysis. OTU at 97% similarity were created, and phylogenetic estimations were performed using Qiime’s (Caporaso et al. 2010) ver. 1.9.0 workflow with no references and all parameters under default conditions. For AOB amoA and AOA amoA, fragment lengths of 500 bp and 130 bp were respectively obtained after merging the corresponding reads and removing the overlapping regions. For these two genes, OTU were created at the 97% similarity threshold using the “usearch” classifier, and the BLAST method (Altschul et al. 1990) was used for assigning taxonomy. All unidentified OTU and those corresponding to the Kingdom Protista were removed before downstream analysis.
Calculations and statistical analysis
All soil and plant data were subjected to statistical analysis using CropStat ver. 7.2 (IRRI, Philippines). Two-way ANOVA was first applied to each sampling period (soil depth × treatment), and when a significant interaction was observed, each soil depth’s data were analyzed by one-way ANOVA. When a significant effect was detected (P < 0.05), the LSD post-hoc tests (P < 0.05) were used to test the differences between treatments. R version 3.4.4 (2018-03-15) was used to generate the figures for potential nitrification. Spearman’s rank correlations between soil properties and gene abundances were calculated in PAST v.2.17 (Hammer et al. 2001). The relative abundances (RA) of OTU generated by Illumina MiSeq were calculated, and Permanova was used to analyze the significance of the OTU’s RA in the different treatments, with 9999 permutations. The “diversity” command of PAST calculated alpha diversity indices, including Shannon’s, inverse (1/D) Simpson’s, Margalef richness, and Chao-1, and generated the rarefaction curves of the different taxa. The average diversity indices for each gene were statistically compared between treatments (Tukey’s LSD test with an alpha of 0.05) using CropStat ver. 7.2 (IRRI, Philippines). The relationships between soil properties and the abundance of AOA, mostly influenced by sorgoleone, were determined using the non-metric multidimensional scaling (NMDS) with the data standardized by the Gower distance.
Results
Chemical properties of soil
The initial soil contained 1147 mg kg−1 soil total P and 25.5 mg kg−1 soil available P (Bray-2 method) and was slightly acidic (pH 5.63). Significant variations in soil pH were observed only at 2.3 months in the 0–10 cm and 10–30 cm depths. In these horizons, the fertilized (120 N) treatments showed lower pH values than 0 N treatments (Table 2). Among fertilized treatments, the no plant control and 296B had, in general, significant lower pH. In the 10–30 cm depth, the pH was not different between the fertilized and non-fertilized 296B, IS20205, and IS32234. The soil extracts collected at 7.5 cm also had significantly lower pH in the control and 296B treatments compared to IS20205 and IS32234 on days 13, 27, 31, and 34 after N supply (Table S1).
The NO3−-N content in soil sampled at 1 and 2.3 months is shown in Table 2. The levels were in general significantly lower in IS20205 and IS32234 in all horizons. At 2.3 months, the topsoil (0–10 cm) of fertilized control contained a very high level of nitrates (22.27 μg g−1 dry soil) compared to the fertilized plants where the highest NO3−-N level was 2.43 μg g−1 dry soil. The nitrate concentrations in the soil extracts collected at 7.5 cm depth every 3–4 days are reported in Table 3 and were also significantly lower in IS20205 and IS32234 compared to 296B and controls. Before day 21, the 0 N and 20 N controls had similar NO3−-N concentrations but significantly higher than that of the other treatments. From day 21, the NO3−-N concentration of the fertilized control increased tremendously due to the supply of other 40 N and 60 N and the absence of plant for absorption. Comparing the NO3−-N level of each treatment in the three horizons, we observed an increasing trend towards deeper horizons.
For NH4+-N, significant differences between treatments were only observed at 1 month. At this sampling period, the rhizosphere of plant treatments contained in general higher amounts of exchangeable NH4+-N than the no plant controls. At the same time, the controls showed significantly lower amounts of total N and total C than plant treatments when no N was applied at either 1 or 2.3 months and regardless to the horizon. Under 20 N (1 month), the control and IS20205 soils contained similar levels of total N in 0–10 cm depth but lower than that of 296B and IS32234. At 2.3 months when 120 N was applied, the fertilized no plant control soils contained significantly less total N and total C although they were estimated roughly. The C/N ratio did not follow a clear pattern and could either be higher or less in the controls.
PNR
Figures for soil PNR were prepared using R software (R Dvelopment Core Team 2018). Two-way ANOVA, carried out between treatments and soil depths, indicated a non-significance of the interaction (P = 0.725) at 1 month, indicating that the trend of PNR variation between treatments was similar in the three depths (Fig. 1a). In each depth, PNR did not significantly differ between treatments (P = 0.499 in 0–10 cm, P = 0.604 in 10–30 cm, P = 0.359 in 30–80 cm) although in 0–10 cm, 296B treatments and the fertilized control showed slightly higher values, and in 10–30 cm and 30–80 cm, PNR in controls was slightly higher. At 2.3 months, the two-way ANOVA revealed a significant interaction between treatments and soil depths (P ≤ 0.001), which indicated that the trend of PNR variation between treatments differed across soil horizons. Globally, significantly (P ≤ 0.05) higher PNR was obtained in the 0–10 cm than in 10–30 cm and 30–80 cm soil depths (Fig. 1b). When PNRs in each soil depth were compared among themselves, we found that in the 0–10 cm horizon, fertilized treatments showed significantly higher PNR than the non-fertilized (P ≤ 0.001). Moreover, IS20205 and IS32234 showed significantly lower PNRs than 296B in the absence of N application, while under 120 N, IS20205 had lower PNR than IS32234 and 296B. No statistically significant difference in PNR between treatments was observed in 0–10 cm (P = 0.134) and 30–80 cm (P = 0.39).
Plant biomass
Plant root biomass was higher in the 0–10 cm depth (Fig. S2) than in the deeper horizons. IS20205 gave higher root dry weight than IS32234 and 296B at 1 month (20 N). At 2.3 months (120 N), IS20205 and IS32234 showed similar root biomass, regardless of the fertilization type and soil depth. The total root biomass at 1 month (Fig. 2a) was significantly higher in IS20205, followed by IS32234 then 296B. However, IS20205 and IS32234 had similar root biomass at 2.3 months and it was higher than that of 296B under 0 N and 120 N fertilizations. The variation in plant shoot dry weight followed the same pattern as the total root dry weight (Fig. 2b).
Sorgoleone production
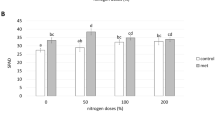
The specific sorgoleone production of the three sorghum lines in each of the three soil depths and the mean specific sorgoleone production in the whole soil column were determined after 1 month and 2.3 months’ growth (Fig. 3). At 1 month, the sorgoleone level of 296B was below the detection limit. IS20205 produced significantly (P ≤ 0.001) higher levels than IS32234 in each soil depth, regardless of the fertilization type. Similarly to 1 month, the level of sorgoleone production of IS20205 at 2.3 months was significantly (P ≤ 0.001) higher than that of IS32234 in the different soil depths. At this period, 296B could release detectable amounts of sorgoleone which were not significantly different from that released by IS32234 in the different soil horizons. The levels of sorgoleone at 2.3 months were 2.18 to 2.50 times that at 1 month for IS20205 and 5 to 10 times that for IS32234. Whether fertilized or not, the sorgoleone production pattern was the same for the three sorghum lines. We observed an increasing trend of sorgoleone levels towards deeper soil horizons for IS20205 during the whole experimental period and for IS32234 at 2.3 months under the N fertilization.
Specific sorgoleone (μg g−1 dry root) of sorghum lines at 1 month and 2.3 months in the soil horizons 0–10 cm, 10–30 cm, and 30–80 cm, without N and with N applications. Bars represent means of 4 replicates ± Stdev. At each sampling period, one-way ANOVA was applied to sorgoleone values in the same horizon and to mean sorgoleone concentration in the 3 horizons. In each horizon and for the means of 3 horizons, bars carrying different letters are significantly different at P < 0.001 Student’s t test. Bdl below detection limit
Microbial communities size as obtained by qPCR
The initial soil contained 3.3 × 108 and 2 × 106 copies g−1 of bacterial and archaeal 16S rRNA genes, respectively. At 1 month, total bacteria varied between 8.2 × 108 and 2 × 109 copies g−1 while total archaeal abundance ranged between 4 × 106 and 1.1 × 107 copies g−1. At 2.3 months, bacterial abundance ranged between 4.5 × 108 and 1.4 × 109 copies g−1 and total archaea varied between 5.8 × 106 and 1.2 × 107 copies g−1. The initial soil harbored 8.8 × 105 and 6.5 × 105 copies g−1 of AOB and AOA amoA genes, respectively. At 1 month, AOB abundance varied between 2.6 × 104 and 1.2 × 105 copies g−1 while AOA ranged between 1.4 × 106 and 5.2 × 106 copies g−1. At 2.3 months, they were estimated to be in the range of 7.2 × 104 and 2.3 × 105 copies g−1 for AOB and between 8.3 × 105 and 9.4 × 106 copies g−1 for AOA (data not shown).
Microbial abundances as expressed in log10 copies per gram of dry soil at 1 month
The variations of microbial abundance at 1 month are shown in Fig. 4. Copy numbers of bacteria, archaea, and AOA in each horizon were significantly (P ≤ 0.005) higher in treatments than in the initial soil. In contrast, the initial soil harbored significantly higher numbers of AOB than treatments in the three horizons (P ≤ 0.001). When treatments were compared among themselves, we observed that bacterial abundance did not significantly differ between treatments in the 0–10 cm depth, but archaea were significantly less abundant (P = 0.002) in fertilized plant treatments than in fertilized control and in non-fertilized IS20205 than in non-fertilized 296B and control. Without N fertilization, abundances of AOA and AOB were not significantly different among treatments. However, AOA were significantly (P = 0.037) lower in the fertilized IS20205 than in the other fertilized treatments, while all fertilized plant treatments harbored similarly but significantly (P = 0.001) lower AOB numbers than the fertilized control. In the 10–30 cm depth, the only significant difference was observed among fertilized treatments for bacteria (P = 0.031) where IS20205 and IS32234 soils harbored higher bacterial abundance than 296B and control. Similarly, IS20205 and IS32234 soils contained significantly (P = 0.031) higher bacterial number in the 30–80 cm depth than 296B and control, and the difference was clearer under 0 N.
Abundances of bacterial and archaeal 16S rRNA genes and archaeal and bacterial amoA genes (AOA-amoA, AOB-amoA) transformed into log10 copies per gram of dry soil in samples taken from 0 to 10 cm, 10 to 30 cm, and 30 to 80 cm horizons of the different treatments after 1-month growth. Initial soil is the same composite sample taken at the beginning of the experiment. Bars represent mean abundances of 4 replicate samples, each measured in quadruplicate qPCR reactions, and respective standard deviations
Microbial abundances as expressed in log10 copies per gram of dry soil at 2.3 months
Microbial abundances at 2.3 months are shown in Fig. 5. Similarly to at 1 month, bacteria, archaea, and AOA in each horizon were significantly (P ≤ 0.001) higher in treatments than in the initial soil. At 2.3 months, the AOB numbers in the initial soil were significantly higher than that in treatments only in the top 10 cm depth (P = 0.037). Differences were statistically significant for bacteria in 0–10 cm (P ≤ 0.001), 10–30 cm (P = 0.002), and 30–80 cm (P ≤ 0.001). In each of these three soil depths, although the bacterial abundances in the N fertilized (120 N) and non-fertilized six plant treatments did not differ among themselves, they were higher than that of the two controls. Here, we observed an increasing trend of the bacterial abundance in the N fertilized compared to the non-fertilized plant treatments. Similarly to bacteria, statistically significant differences were observed for AOA in the three depths at a probability level of P ≤ 0.001 in each depth. In 0–10 cm without N fertilization, IS20205 and IS32234 grouped and contained lower AOA than 296B and the control. The application of 120 N increased the AOA abundance in all treatments, but IS20205 maintained significantly lower levels than IS32234 and 296B, which also contained lower AOA abundance than the fertilized control. In 10–30 cm and 30–80 cm, the distribution of AOA abundance between treatments followed almost the same trend as in 0–10 cm with the difference that under 120 N, there was no difference in AOA abundance between IS20205 and IS32234. Moreover, the application of 120 N did not increase the AOA abundance in these deeper horizons as it was the case in the 0–10 cm depth. On the other hand, statistically significant differences between treatments were not observed for archaea (P = 0.068 in 0–10 cm, P = 0.058 in 10–30 cm, P = 0.112 in 30–80 cm) and AOB (P = 0.13 in 0–10 cm, P = 0.353 in 10–30 cm, P = 0.558 in 30–80 cm) at 2.3 months. However, when the four treatments of each fertilization type were analyzed separately, archaea in the non-fertilized treatments were significantly higher in 296B than in control, IS20205, and IS32234 in 10–30 cm (P = 0.007) and 30–80 cm (P = 0.002).
Abundances of bacterial and archaeal 16S rRNA genes and archaeal and bacterial amoA genes (AOA-amoA, AOB-amoA) transformed into log10 copies per gram of dry soil in samples taken from 0 to 10 cm, 10 to 30 cm, and 30 to 80 cm horizons of the different treatments after 2.3 months growth. Initial soil is the same composite sample taken at the beginning of the experiment. Bars represent mean abundances of 4 replicate samples, each measured in quadruplicate qPCR reactions, and respective standard deviations
Relationships between soil properies and ammonia oxidizer abundance
A Spearman’s correlation was run considering all data from the 1-month and 2.3-month samples for each variable to determine the relationships among soil pH, %moisture, PNR, NO3−-N, NH4+-N, sorgoleone release, and the abundances of AOB and AOA total bacteria and total archaea (Table 4). A weak but positive monotonic correlation was observed between pH and sorgoleone (rs = 0.185) and pH and total bacteria (rs = 0.250), while a weak, negative correlation related the pH and PNR (rs = − 0.196). The soil %moisture influenced most of the parameters but at different strengths. It was positively moderately correlated with the abundance of archaea (rs = 0.498) and with NO3−-N content (rs = 0.481), positively weakly correlated with the abundance of AOB (rs = 0.356) and PNR (rs = 0.246), and positively very weakly correlated with abundance of AOA (rs = 0.161). In contrast, soil %moisture was negatively weakly correlated with bacterial abundance (rs = − 0.357) and negatively moderately correlated with sorgoleone content (rs = − 0.444). Apart from its correlation with %moisture, PNR was positively very weakly correlated with AOA (rs = 0178), positively weakly correlated with AOB (rs = 0.4283) and total archaea (rs = 0.301), and negatively very weakly correlated with exchangeable NH4+-N (rs = − 0.147) and sorgoleone content (rs = − 0.165). NO3−-N content was very weakly, positively correlated with exchangeable NH4+-N content (rs = 0.183); weakly, positively correlated with total archaea (rs = 0.348); but strongly, negatively correlated with sorgoleone content (rs = − 0.611) and strongly positively with AOA abundance (rs = 0.602). Adding to its correlations with PNR and NO3−-N content, exchangeable NH4+-N content was also positively weakly correlated with sorgoleone content (rs = 0.290) and bacterial abundance (rs = 0.320), but negatively weakly with AOB abundance (rs = − 0.207). Sorgoleone content positively moderately correlated with bacterial abundance (rs = 0.407) and negatively moderately with AOA abundance (rs = − 0.489). Furthermore, a weak and negative correlation was observed between sorgoleone content and total archaea abundance (rs = − 0.247). The total archaea abundance positively moderately correlated with AOA abundance (rs = 0.407).
Microbial community composition as obtained by Illumina MiSeq
The rarefaction curves (Fig. S3) showed that our sequencing depth captured well the microbial community diversity.
16S rRNA V4 sequence data
A total of 1,448,418 high-quality 16S rRNA V4 sequences were obtained after chimera removal. The bacterial 16S rRNA gene comprised 1,315,728 sequences with an average of 27,411 reads (18,886–34,348 reads) per sample. They clustered into 27,512 OTU (97% similarily), and several phyla and Permanova revealed significant differences (P < 0.001) between treatments based on OTU-level abundances (data not shown). The dominating 20 phyla with at least 0.1% OTU abundance in at least one of the samples were Acidobacteria, Actinobacteria, AD3, Armatimonadetes, Bacteroidetes, Chlamydiae, Chloroflexi, Cyanobacteria, Elusimicrobia, Firmicutes, GAL15, Gemmatimonadetes, Nitrospirae, OD1, Planctomycetes, Proteobacteria, Tenericutes, Verrucomicrobia, WPS-2, and WS3 (Fig. 6). At 1 month, Acidobacteria, Nitrospirae, Gemmatinomonadetes, Elusimimicrobia, Planctomycetes, and Verrumicrobia significantly dominated in controls than in plant treatments with P ≤ 0.001 for all the named phyla except Verrumicrobia (P = 0.009). No statistical significance (P = 0.073) was observed for Proteobacteria at this period. The number of Actinobacteria significantly (P ≤ 0.001) increased in plant treatments under N application at 1 month, and the increase was stronger in IS20205. At 2.3 months, Acidobacteria did not show a statistical difference under 0 N, but under 120 N, plant treatments showed significantly (P ≤ 0.001) lower abundance than the control. As at 1 month, a significantly lower abundance of Nitrospirae (P ≤ 0.001) was obtained at 2.3 months. In contrast, in this period, Proteobacteria were significantly more abundant in plant treatments than in controls (P ≤ 0.001). At 2.3 months, Actinobacteria abundance was significantly (P ≤ 0.001) higher in plant treatments than in controls, especially under 120 N. Alpha diversity indices generated by PAST and subjected to 2-way ANOVA (treatment × sampling period) showed significant probability levels for the Menhinick and Margalef indices (Table S2). With 0 N, these indices were significantly lower in IS20205 than in the other treatments. With the N application, they were still lower in IS20205, but the differences with the other treatments were not significant.
The archaeal 16S rRNA gene comprised 65,768 sequences with an average of 1370 reads (345–4359 reads) per sample and clustered into 488 OTU. Methanomassiliicoccaceae, Nitrososphaeraceae, and unclassified NRP-J were the most abundant families (Fig. 7). The unclassified NRP-J was generally higher under 0 N. In contrast, N application increased Nitrososphaeraceae abundance as we observed at 2.3 months. Significant differences between treatments were observed for Taxa-S, Individuals, Shannon, Brouillon, Menhinick, and Margalef indices (Table S3). These indices were higher in controls than in fertilized and non-fertilized plant treatments. Plant treatments in each fertilization type did not exhibit significant differences among them for Taxa_S, Individuals, and Menhinick. However, fertilized plant treatments presented lower diversities than the non-fertilized. The Shannon index was significantly lower in IS20205 than in 296B and control under 0 N, and under N fertilization, it was significantly lower in IS20205 compared to IS32234.
Relative abundance of 16S rRNA Archaea based on family OTU reads at 1 and 2.3 months. The 7 most abundant families (i.e., at least 0.1% abundance in at least one of the samples) were used. They represent 99.86 to 100% of OTU relative abundance in the different these treatments. Eury Euryarchaeota, Cren Crenarchaeota, Parv_Parv Parvarchaeota_Parvarchaea (Phylum_Class), Thermo Thermoplasmata, Thaum Thaumarchaeota, Unass and Un Unassigned, Cenarch Cenarchaeales, Methanom Methanomassiliicoccaceae, Nitros_Nitros Nitrososphaerales_Nitrososphareaceae
AOA and AOB amoA sequence data
Illumina MiSeq generated 343,910 AOA amoA sequences (77.25% of the raw sequences) after removing no-hits and sequences of less than 97% similarity with amoA references. These sequences averaging 14,330 reads per sample clustered into 120 OTU. The eight most abundant OTU (1, 2, 3, 4, 6, 7, 8, 9) with at least 2% abundance in at least one of the samples are shown in Fig. S4. These OTU were identified (100% similarity) as amoA genes of uncultured soil archaeon clones. Permanova revealed significant differences (P < 0.001) between treatments (data not shown). N fertilization increased the relative abundance of OTU_1 and OTU_7 but decreased that of OTU_2, OTU_3, OTU_4, OTU_6, and OTU_8. Without N, the relative abundance of OTU_1 was significantly lower in control than in plant treatments, but under 120 N, the difference was not marked. It contrasted with the relative abundance of OTU_2, which was significantly lower in the fertilized control. Without N, the relative abundance of OTU_3 was lower in IS20205 than in the other treatments, but under 120 N, all plant treatments had a lower relative abundance of OTU_3 than the control. The other relative abundance of OTU also showed significant differences between treatments. Alpha diversity indices were not different between treatments (data not shown). The NMDS ordination revealed a clear separation of AOA as influenced by the fertilization type (Fig. S5). Plants treatments tightly grouped and were separated with controls, especially under 120 N. NO3 and NO2 (potential nitrification) were well influenced by N fertilization and sorgoleone content correlated well with IS20205 and IS32234.
AOB amoA MiSeq showed that only 24.59% of the filtered reads were merged, and only three OTU were 97% similar to closest AOB references. Therefore, AOB MiSeq data were omitted for downstream analysis.
Discussion
High nitrification has been a concern for decades for the negative impact on the environment, due to high N losses via nitrate leaching which cause soil pollution or emission of nitrous oxide (N2O) through denitrification (Timilsena et al. 2015; Uchida and von Rein 2018). Reducing nitrification rates in soils represents a strategy to mitigate the negative impact of nitrate to the environment (Subbarao et al. 2012). Biological nitrification inhibition (BNI) is a plant-mediated phenomenon that inhibits nitrification by specific secondary metabolites exuded from roots (Sorgoleone, Brachialactone, Sakuranetin, 1,9-Decanediol) and appears to be an alternative to the costly chemical fertilizers (Subbarao et al. 2015). Sorgoleone, a known sorghum metabolite, was identified as an essential BNI component of hydrophobic-BNI activity of exudates from sorghum roots which can potentially reduce the abundance of AOB and AOA amoA genes (Coskun et al. 2017; Subbarao et al. 2013; Dayan et al. 2010). However, few studies have thoroughly examined the relationships between sorgoleone release, environmental factors, potential nitrification, and composition and abundance of microbes, including AOB and AOA amoA genes. Here, we investigated such relationships in the presence of the growth of three sorghum lines with different sorgoleone production abilities. The potential nitrification rate (PNR) was generally found lower in the rhizosphere of the higher sorgoleone producing genetic stock IS20205, more specifically in the top 10 cm (Fig. 1). This lower PNR under the higher sorgoleone line may partly explain the lower nitrate concentration in the soil extracts collected at 7.5 cm of this line. Furthermore, since IS20205 and IS32234 showed similar root and shoot biomass, particularly at 2.3 months, they likely absorbed soil nutrients in the same manner. Therefore, the difference the difference in sorgoleone release could largely explain the difference in NO3−-N concentration in soil solutions. At times, low nitrification rates have been explained by a decline in NH4+ supply rather than the toxicity of specific compounds to nitrifiers (Schimel et al. 1996). In this study, the application of 120 N increased the exchangeable NH4+-N content, which correlated with higher potential nitrification. However, Spearman’s correlations reinforced that sorgoleone plays an integral part in the nitrification inhibition of sorghum. Our data showed that higher sorgoleone release led to stronger inhibition of AOA and potential nitrification and lower nitrate content in the soil. In contrast, differences in sorgoleone levels between sorghum lines did not influence the abundance of AOB, although plant treatments contained less AOB than controls. It has been recognized that AOA outnumber AOB in many habitats (Rughöft et al. 2016; Herrmann et al. 2012) and can play a dominant role in nitrification, particularly in acid soils (Prosser and Nicols 2012; Hu et al. 2014). They also can thrive at much lower substrate concentration than AOB due to higher substrate affinity (Zhang et al. 2012; Yao et al. 2013). The acidic condition and low exchangeable NH4+-N and NO3−-N concentrations of the soil used in this study (Table 2) may favor the dominance of AOA over AOB.
Although AOB were not identified clearly in this study by amoA amplicon sequencing, their abundance obtained by qPCR was positively correlated with soil moisture and potential nitrification, thus supporting the well-known contribution of AOB to soil nitrification (Jia and Conrad 2009). Frequently, it has been reported a positive relationship between AOA abundance and soil nitrification potential (Yao et al. 2011) or nitrate production (Offre et al. 2009; Gubry-Rangin et al. 2010). Gao et al. (2016) demonstrated that AOA accounted for most of the ammonia oxidation in fine particulate matter, with a low contribution of AOB, and this was justified by the higher abundance of AOA in samples. Zhang et al. (2012) also reported a positive correlation between AOA, but not AOB, and soil nitrate concentration. The predominance of AOA in many environments compared to AOB justified that the ammonia oxidation process promoted by AOA may be more intense (Jin et al. 2011; Hou et al. 2013). Consistent with these previous studies, the higher abundance of AOA over AOB may indicate that AOA mostly mediate the ammonia oxidation in the present work. The negative correlation between AOA abundance and sorgoleone content and the lack of correlation between the metabolite and AOB abundance clearly showed that the biological control of nitrification in sorghum cultivation is mostly related to the inhibition of AOA by sorgoleone than AOB. However, the abundance of the microbial population does not necessarily directly link to enzymatic activity. Therefore, since we did not extend the analysis to mRNA or protein levels, we cannot exclude the possibility that AOB contributed similarly or much more than AOA. Sterngren et al. (2015) stated that AOA are important drivers of nitrification under nitrogen-poor conditions, but high available N results in increased abundance, activity, and relative importance of AOB for gross nitrification in grassland soil. Besides, Fu et al. (2018) demonstrated the importance of AOB nitrification after N application to agricultural soils in a long-term field experiment and reported the inhibitory effect of some chemical inhibitors including DCD on the abundance and activity of AOB.
On the other hand, bacteria outnumbered archaea in this study. This result confirmed most reports that showed bacteria dominate archaea in multiple environments (Timonen and Bomberg 2009; Bates et al. 2011; Chen et al. 2017). Illumina MiSeq data showed that the majority of archaeal 16S rRNA sequence reads in the 0–10 cm soil layer belonged to the Thaumarchaeotal class. More specifically, they belonged to the ammonia-oxidizing genus Candidatus Nitrosphaera (Nitrososphaeraceae family) or other unclassified genera of the order Cenarchaeales, which dominated the archaeal community across all treatments (Fig. 7). Our data agree with the information available on archaeal communities in other soils, i.e., a general dominance of the Crenarchaeota (Pester et al. 2011), along with minor Euryarchaeota (Bates et al. 2011) and Parvarchaeota. Here, the dominance of Nitrososphaeraceae increased with soil acidity due to the overall application of 120 N, as shown in Table 2 and Fig. 7. This result indicates that this group of ammonia-oxidizing archaea is regulated (i.e., decreased) by increasing pH, as reported in forest soils (Lehtovirta et al. 2009). Since Thaumarchaeota known to harbor most AOA dominated the archeal 16S rRNA, any variation in the abundance and composition of AOA would lead to a similar variation in that of total archaea. In this regard, we found that the abundance of both archaeal amoA and 16S rRNA genes positively correlated with soil moisture, potential nitrification, NO3−-N content, and the abundance of the two genes were positively related each other as shown by Spearman’s correlations (Table 5). Moreover, the abundance of the two genes was both negatively correlated with the amount of sorgoleone. Higher release of sorgoleone lowered the abundance of both genes and decreased the nitrification.
The abundance of total bacteria, as revealed by qPCR, was not significantly different between treatments at 1 month where half of the treatments received only 20 N. However, at the late growth stage and with the application of 120 N, total bacteria were more abundant in plant treatments than in controls. Furthermore, bacteria were also less abundant in the initial soil. These results indicated that under favorable conditions, the bacterial abundance increases. In this regard, Spearman’s correlations had shown a positive correlation between abundances of bacteria and exchangeable NH4+-N. In addition to the exchangeable NH4+-N content, total C and total N were significantly higher in the plant treatments than in controls in each of the three horizons (Table 2). This result agrees with findings from other studies reporting that the abundance of heterotrophic bacteria increases under high nutrient availability (Leff et al. 2015; Gómez-Pereira et al. 2012). Marschner (2011) reported that roots could release as much as 40% of net photosynthetic C in the rhizosphere, and it includes compounds such as amino acids, organic acids, sugars, phenolics and other secondary metabolites (Bertin et al. 2003). On the other hand, our MiSeq data (Fig. 6) showed that some bacterial phyla increased with increasing soil nutrient content, while others decreased. It is consistent with Rughöft et al. (2016) who reported that nutrient availability also drives the bacterial community composition. Moreover, these results agree well with the copiotrophic hypothesis (Fierer et al. 2007), which predicts that fast-growing copiotrophic soil bacteria such as Proteobacteria, Actinobacteria, and Bacteroidetes prefer nutrient-rich environments, while slow-growing oligotrophs (e.g., Acidobacteria, Nitrospirae) would thrive in oligotrophic soils.
We found that the abundance of bacteria did not correlate with that of AOB, unlike the abundances of total archaea and AOA. The very low abundance of AOB compared to total bacteria as obtained by qPCR may justify this lack of correlation. However, bacterial abundances determined by qPCR could be slightly biased since the numbers of ribosomes can vary among bacteria. Furthermore, the Illumina MiSeq detected shallow AOB amoA sequence reads. Only 24.59% of the filtered AOB amoA reads could merge, and more than half of the merged OTU were unclassified. Also, the sequence similarity between the identified OTU and reference AOB amoA genes varied between 75 and 100%. Each molecular method may come alone with a bias, and we cannot rule out that our AOB community composition may have been influenced by the PCR conditions or the set of primer used. However, some studies that have used the same AOB primer set (Meinhardt et al. 2015; Gao et al. 2016) have previously reported a high recovery of soil AOB composition, including clearly identified Beta-Proteobacterial genera harboring the amoA gene.
In the present study, the sorghum genetic stock IS20205 which releases higher amounts of sorgoleone per gram of dry root showed lower diversity and less abundance of AOA in its rhizosphere than in the rhizosphere soils of the other two lines. This sorghum line developed higher root and shoot biomass compared to the medium sorgoleone-producing line IS32234 and the lower sorgoleone-producing line 296B in the early growth stage when no or little amount of N as 20 N was applied. During this period where soil N was low, the better nitrification control by IS20205 may have provided the plant with more exchangeable NH4+-N for uptake and its higher roots may allow a more efficient uptake of the formed NO3−-N compared to the 2 other genetic stocks. Consequently, better uptake could lower the nutrient content in the rhizosphere of IS20205 (Table 2), thus leading to higher root development. At 2.3 months, IS20205 and IS32234 showed similar root and shoot biomass, which was significantly higher than that of 296B. The high and medium sorgoleone-producing lines may absorb soil nutrients similarily during the later growth stage. Therefore, the differences observed in potential nitrification rates and the composition of nitrifiers evidently arose from the difference in sorgoleone release. The positive effect of sorgoleone on nitrification inhibition and plant growth indicates that it does not induce the overall plant-microbial interactions negatively as it was observed in cucumber by the autotoxin metabolite p-Coumaric (Zhou et al. 2018).
Conclusion
To our knowledge, this is the first study investigating in detail the BNI function in sorghum as influenced by differences in sorgoleone release and environmental factors. It demonstrated that the concentration of sorgoleone released by sorghum roots is an active driver of soil nitrifier community composition and has a stronger influence on AOA. Sorghum plants that exude higher specific sorgoleone (μg g−1 dry root) exhibit more significant inhibition of the abundance of AOA amoA genes, slowing down soil nitrification and indirectly reducing the emission of nitrous oxide (N2O). Other environmental factors such as soil moisture, nutrient contents, and pH to a lower extent also influenced the AOA community composition and distribution, as well as that of total bacteria, total archaea, and AOB. The abundances of total bacteria, archaea, AOA, and AOB amoA genes also varied across soil depths and sorghum growth stages. Overall, the study showed that sorghum lines with higher sorgoleone-release potential have better control of soil nitrification, mainly by inhibiting the abundance of AOA amoA genes, and the subsequent biological nitrification inhibition contributed for better plant growth.
Change history
23 December 2019
The name of the author was incorrectly spelled as “Filippo Carannant”. The correct spelling is “Filippo Carannante” and is now presented correctly in this article.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Apprill A, McNally S, Parsons R, Weber L (2015) Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol 75:129–137. https://doi.org/10.3354/ame01753
Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N (2011) Examining the global distribution of dominant archaeal populations in soil. ISME J 5:908–917. https://doi.org/10.1038/ismej.2010.171
Belser LW, Mays EL (1980) Specific inhibition of nitrite oxidation by chlorate and its use in assessing nitrification in soils and sediments. Appl Envrion Microbiol 39:505–510
Bertin C, Yang XH, Weston LA (2003) The role of root exudates and allelochemicals in the rhizosphere. Plant Soil 256:67–83
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peňa AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Chen X, Andersen TJ, Morono Y, Inagaki F, Jorgensen BB, Lever MA (2017) Bioturbation as a key driver behind the dominance of bacteria over archaea in near-surface sediment. Sci Rep 7:2400. https://doi.org/10.1038/s41598-017-02295-x
Coskun D, Britto DT, Shi W, Kronzucker HJ (2017) Nitrogen transformations in modern agriculture and the role of biological nitrification inhibition. Nat Plants 3:17074. https://doi.org/10.1038/nplants.2017.74
Czarnota MA, Rimando AM, Weston LA (2003) Evaluation of seven sorghum (Sorghum sp.) accessions. J Chem Ecol 29:2073–2083
Dayan FE, Rimando AM, Pan Z, Baerson SR, Gimsing AL, Duke SO (2010) Sorgoleone. Phytochem 10:1032–1039. https://doi.org/10.1016/j.phytochem.2010.03.011
DeLong EF (1992) Archaea in coastal marine environments. Proc Natl Acad Sci U S A 89:5685–5689. https://doi.org/10.1073/pnas.89.12.5685
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinf 26:2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Einhellig FA, Souza IF (1992) Phytotoxicity of sorgoleone found in grain sorghum root exudates. J Chem Ecol 18:1–11
Fierer N, Bradford MA, Jackson RB (2007) Toward and ecological classification of soil bacteria. Ecol 88:1354–1364. https://doi.org/10.1890/05-1839
Forster JC (1995) Soil nitrogen. In: Alef K, Nannipieri P (eds) Methods in applied soil microbiology and biochemistry. Academic Press, San Diego, pp 79–87
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonoa-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A 102:14683–14688
Fu Q, Clark IM, Zhu J, Hu H, Hirsch PR (2018) The short-term effects of nitrification inhibitors on the abundance and expression of ammonia and nitric oxidizers in a long-term field experiment comparing land management. Biol Fertil Soils 54:163–172. https://doi.org/10.1007/s00374-017-1249-2
Gao JF, Fan XY, Pan KL, Li HY, Sun LX (2016) Diversity, abundance and activity of ammonia-oxidizing microorganisms in fine particulate matter. Sci Rep 6:38785. https://doi.org/10.1038/srep38785
Gómez-Pereira PR, Schuler M, Fuchs BM, Bennke C, Teeling H, Waldmann J, Richter M, Barve V, Bataille E, Glöckner FO, Amann R (2012) Genomic content of uncultured Bacteroidetes from contrasting oceanic provinces in North Atlantic Ocean. Environ Microbiol 14:52–66
Gubry-Rangin C, Nicol GW, Prosser JI (2010) Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol Ecol 74:566–574. https://doi.org/10.1111/j.1574-6941.2010.00971.x
Hammer O, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron:4(1), 9pp
Hannon GJ (2010) FASTX-toolkit. http://hannonlab.cshl.edu/fastx_toolkit. Accessed 22 Aug 2019
Hayatsu M, Tago K, Saito M (2008) Various players in the nitrogen cycle: diversity and functions of the microorganisms involved in nitrification and denitrification. Soil Sci Plant Nutr 54:33–45
Herrmann M, Hädrich A, Küsel K (2012) Predominance of Thaumarchaeal ammonia oxidizer abundance and transcriptional activity in an acid fen. Environ Microbial 14:3013–3025. https://doi.org/10.1111/j.1462-2920.2012.02882.x
Hou J, Cao XY, Song CL, Zhou YY (2013) Predominance of ammonia-oxidizing archaea and nirK-gene-bearing denitrifiers among ammonia-oxidzing and denitrifying populations in sediments of a large urban eutrophic lake (lake Donghu). Can J Microbiol 59:456–464. https://doi.org/10.1139/cjm-2013-0083
Hu HW, Xu ZH, He JZ (2014) Ammonia-oxidizing archaea play a predominant role in acid soil nitrification. Adv Agron 125:261–302
Iizumi T, Mizumoto M, Nakamura KA (1998) Bioluminescence assay using Nitrosomonas europaea for rapid and sensitive detection of nitrification inhibitors. Appl Environ Microbiol 64:3656–3662
Ishikawa T, Subbarao GV, Ito O, Okada K (2003) Suppression of nitrification and nitrous oxide emission by tropical grass Brachiaria humidicola. Plant Soil 255:413–419
Jia ZJ, Conrad R (2009) Bacteria rather than archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol 11:1658–1671. https://doi.org/10.1111/j.1462-2920.2009.01891.x
Jin T, Zhang T, Ye L, Yee OO, Wong YH, Qian PY (2011) Diversity and quantity of ammonia-oxidizing archaea and bacteria in sediment of the Pearl River estuary, China. Appl Microbiol Biotechnol 90:1137–1145. https://doi.org/10.1007/s00253-011-3107-8
Jones N (2013) Grass gets greener. Nature 501:291
Klotz MG, Alzerreca J, Norton JM (1997) A gene encoding a membrane protein exists upstream of the amoA/amoB genes in ammonia-oxidizing bacteria; a third member of the amo operon? FEMS Microbiol Lett 150:65–73
Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA (2005) Isolation of an autotrophic ammnonia-oxidizing marine archaeon. Nature 437:543–546
Leff JW, Jones SE, Prober SM, Barberán A, Borer ET, Firn JL et al (2015) Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc Natl Acad Sci U S A 112:10967–10972. https://doi.org/10.1073/pnas.1508382112
Lehtovirta LE, Prosser JI, Nicol GW (2009) Soil pH regulates the abundance and diversity of group 1.1c Crenarchaeota. FEMS Microbiol. Ecol 70:367–378. https://doi.org/10.1111/j.1574-6941.2009.00748.x
Li M, Wei G, Shi W, Sun Z, Li H, Wang X, Gao Z (2018) Distinct distribution patters of ammonia-oxidizing archaea and bacteria in sediment and water column of the yellow river estuary. Sci Rep 8:1584. https://doi.org/10.1038/s41598-018-20044-6
Loy A, Lehner A, Lee N, Adamczyk J, Meier H, Ernst J, Schleifer KH, Wagner M (2002) Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl Environ Microbiol 68:5064–5081. https://doi.org/10.1128/AEM.68.10.5064-5081.2002
Magoc T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. https://doi.org/10.1093/bioinformatics/btr507
Marschner H (2011) Marchner’s mineral nutrition of higher plants, 3rd edn. Academic Press, London 672pp
Meinhardt KA, Bertagnolli A, Pannu MW, Strand SE, Brown SL, Stahl DA (2015) Evaluation of revised polymerase chain reaction primers for more inclusive quantification of ammonia-oxidizing archaea and bacteria. Environ Microbiol Rep 7:354–363. https://doi.org/10.1111/1758-2229.12259
Morimoto S, Hayatsu M, Takada-Hoshino Y, Nagaoka K, Yamazaki M, Karasawa T, Takenaka M, Akiyama H (2011) Quantitative analyses of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) in fields with different soil types. Microbes Environ 26:248–253
Nardi P, Akutsu M, Pariasca JT, Wissuwa M (2013) Effect of methyl 3-4-hydroxyphenyl propionate, sorghum root exudate, on N dynamic, potential nitrification activity and abundance of ammonia-oxidizing bacteria and archaea. Plant Soil 367:627–637
Nercessian O, Fouquet Y, Pierre C, Prieur D, Jeanthon C (2005) Diversity of bacteria and archaea associated with a carbonate-rich metalliferous sediment sample from the rainbow vent field on the Mid-Atlantic Ridge. Environ Microbiol 7:698–714
Netzly DH, Buler LG (1986) Roots of sorghum exudate hydrophobic droplets containing biologically active components. Crop Sci 26:775–778
Offre P, Prosser JI, Nicol GW (2009) Growth of ammonia-oxidizing archaea in soil microcosms is inhibited by acetylene. FEMS Microbiol Ecol 70:99–108. https://doi.org/10.1111/j.1574-6941.2009.00725.x
Okano Y, Hristova KR, Leutenegger CM, Jackson LE, Denison RF, Gebreyesus B, Lebauer D, Scow KM (2004) Application of real-time PCR to study effeccts of ammonium on populationsize of ammonia-oxidizing bacteria in soil. Appl Environ Microbiol 70:1008–1016
Parada AE, Needham DM, Fuhrman JA (2016) Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18(5):1403–1414. https://doi.org/10.1111/1462-2920.13023
Pester M, Schleper C, Wagner M (2011) The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr Opin Microbiol 14:300–306. https://doi.org/10.1016/j.mib.2011.04.007
Prasad R, Power PJ (1995) Nitrification inhibitors for the agriculture health and environment. Adv Agron 54:233–281
Prosser JI, Nicol GW (2012) Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol 20:523–531. https://doi.org/10.1016/j.tim.2012.08.001
Purkhold U, Pommerening-Roser A, Juretschko S, Schmid MC, Koops HP, Wagner M (2000) Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implication for molecular diversity surveys. Appl Environ Microbiol 66:5368–5382
R Development Core Team (2018) R: a language and environmental for statistical computing. R Foundation for Statistical Computing, Vienna https://www.R-project.org. Accessed 27 May 2019
Raun WR, Johnson GV (1999) Improving nitrogen use efficiency for cereal production. Agron 54:73–83
Rughöft S, Hermann M, Lzar CS, Cesarz S, Levick SR, Trumbore SE, Küsel K (2016) Community composition and abundance of bacterial, archaeal and nitrifiying populations in savanna soils on contrasting pedrock material in Kruger national park, South Africa. Front Microbiol 7:1638. https://doi.org/10.3389/fmicb.2016.01638
Sarr PS, Sugiyama A, Begoude DAB, Yazaki K, Araki S, Nawata E (2019) Diversity and distribution of arbuscular mycorrhizal fungi in cassava (Manihot esculenta Crantz) croplands in Cameroon as revealed by Illumina MiSeq. Rhiz. https://doi.org/10.1016/j.rhisph.2019.100147
Schimel JP, Van Cleve K, Cates RG, Clausen TP, Reichardt PB (1996) Effects of balsam poplar (Populus balsmifera) tannins and low molecular weight phenolics on microbial activity in taiga floodplain soil: implication for changes in N cycling during succession. Can J Bot 74:84–90
Schlesinger WH (2009) On the fate of anthropogenic nitrogen. Proc Natl Acad Sci U S A 106:203–208
Schöler A, Jacquiod S, Vestergaard G, Schulz S, Schloter M (2017) Analysis of microbial communities based on amplicon sequencing of marker genes. Biol Fertil Soils 53:485–489. https://doi.org/10.1007/s00374-017-1205-1
Smith CW, Frederiksen RA (2000) Sorghum: origin, history, technology, and production. Willey, New York
Sterngren AE, Hallin S, Bengtson P (2015) Archaeal ammonia oxidizers dominate in numbers, but bacteria drive gross nitrification in N-amended grassland soil. Front Microbiol 6:1350. https://doi.org/10.3389/fmicb.2015.01350
Subbarao GV, Ito O, Sahrawat KL, Ishikawa T, Berry WL, Nakahara K, Ishikawa T, Watanabe T, Suenaga K, Rondon M, Rao IM (2006a) Scope and strategies for regulation of nitrification in agricultural systems-challenges and opportunities. Crit Rev Plant Sci 25:303–335
Subbarao GV, Ishikawa T, Ito O, Nakahara K, Wang HY, Berry WL (2006b) A bioluminescence assay to detect nitrification inhibitors released from plant roots: a case study with Brachiaria humidicola. Plant Soil 25(1–2):303–335. https://doi.org/10.1007/s11104-006-9094-3
Subbarao GV, Rondon M, Ito O, Ishikawa T, Rao IM, Nakahara K, Lascano C, Berry WL (2007) Biological nitrification inhibition (BNI)-is it a widespread phenomenon? Plant Soil 294:5–18. https://doi.org/10.1007/s11104-006-9094-3
Subbarao GV, Nakahara K, Hurtado MP, Ono H, Moreta DE, Salcedo AF, Yoshihashi T, Ishikawa T, Ishitani M, Ohnishi-Kameyama M, Yoshida M, Rondon M, Rao IM, Lascano CE, Berry WL, Ito O (2009) Evidence for biological nitrification in Brachiaria pastures. Proc Natl Acad Sci U S A 106:17302–17307
Subbarao GV, Sahrawat KL, Nakahara K, Ishikawa T, Kudo N, Kishii M, Rao IM, Hash CT, George TS, Srinivasa RP, Nardi P, Bonnett D, Berry WL, Suenaga K, Ito O, Lata JC (2012) Biological nitrification inhibition (BNI) - a novel strategy to regulate nitrification in agricultural systems. Adv Agron 114:249–302
Subbarao GV, Nakahara K, Ishikawa T, Ono H, Yoshida M, Yoshihashi T, Zhu Y, Zakir HAKM, Deshpande SP, Hash CT, Sahrawat KL (2013) Biological nitrification inhibition (BNI) activity in sorghum and its characterization. Plant Soil 366:243–259
Subbarao GV, Yoshihashi T, Worthington M, Nakahara K, Ando Y, Sahrawat KL, Rao IM, Lata JC, Kishii M, Braun HJ (2015) Suppression of soil nitrification by plants. Plant Sci 233:155–164. https://doi.org/10.1016/j.plantsci.2015.01.012
Subudhi PK, Nguyen HT (2000) Linkage group alignment of sorghum RFLP maps using a RIL mapping population. Genome 43:240–249
Takada-Hoshino Y, Matsumoto N (2004) An improved DNA extraction method using skim milk from soils that strongly adsorb DNA. Microbes Environ 19:13–19
Takada-Hoshino Y, Matsumoto N (2005) Skim milk drastically improves the efficacy of DNA extraction from Andosol, a volvanic ash soil. JARQ 39:247–252
Takai K, Horikoshi K (2000) Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl Environ Microbiol 66:5066–5072. https://doi.org/10.1128/AEM.66.11.5066-5072.2000
Tesfamariam T, Yoshinaga H, Deshpande SP, Rao PS, Sahrawat KL, Ando Y, Nakahara K, Hash CT, Subbarao GV (2014) Biological nitrification inhibition in sorghum: the role of sorgoleone production. Plant Soil. https://doi.org/10.1007/s11104-014-2075-z
Timilsena YP, Adhikari R, Casey P, Muster T, Gill H, Adhikari B (2015) Enhanced efficiency fertilisers: a review of formulation and nutrient release patterns. J Sci Food Agric 95:1131–1142
Timonen S, Bomberg M (2009) Archaea in dry soil environments. Phytochem Rev 8:505–518. https://doi.org/10.1007/s11101-009-9137-5
Treusch AH, Leininger S, Kletzin A, Schuster SC, Klenk HP, Schleper C (2005) Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol 7:1985–1995
Uchida Y, von Rein I (2018) Mitigation of nitrous oxide emissions during nitrification and denitrification processes in agricultural soils using enhanced efficiency fertilizers. In: Vázquez-Luna D, del Carmen M (Eds) Soil contamination and alternatives for sustainable development. IntechOpen. https://doi.org/10.5772/intechopen.81548
Vestergaard G, Schulz S, Schöler A, Schloter M (2017) Making big data smart-how to use metagenomics to understand soil quality. Biol Fertil Soils 53:479–484. https://doi.org/10.1007/s00374-017-1191-3
Yao H, Campbell CD, Chapman SJ, Freitag TE, Nicol GW, Singh BK (2013) Multi-factorial drivers of ammonia oxidizer communities: evidence from a national soil survey. Environ Microbiol 15:2545–2556
Yao H, Gao Y, Nicol GW, Campbell CD, Prosser JI, Zhang I, Han W, Singh BK (2011) Links between ammonia oxidizer community structure, abundance, and nitrification potential in acidic soils. Appl Environ Microbiol 77:4618–4625. https://doi.org/10.1128/AEM.00136-11
Zakir HAKM, Subbarao GV, Pearse SJ, Gopalakrishnan S, Ito O, Ishikawa T, Kawano N, Nakahara K, Yoshihashi T, Ono H, Yoshida M (2008) Detection, isolation and characterization of a root-exudate compound, methyl 3-(4-hydroxy-phenyl) propionate, responsible for biological nitrification inhibition by sorghum (Sorghum bicolor). New Phytol 180:442–451
Zhang LM, Hu HW, Shen JP, He JZ (2012) Ammonia oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acid soils. ISME J 6:1032–1045. https://doi.org/10.1038/ismej.2011.168
Zhou X, Zhang J, Pan D, Ge X, Jin X, Chen S, Wu F (2018) p-Coumaric can alter the composition of cucumber rhizosphere microbial communities and induce negative plant-microbial interactions. Biol Fertil Soil 54:363–372. https://doi.org/10.1007/s00374-018-1265-x
Acknowledgments
This study was implemented under the Japan International Research Center for Agricultural Sciences (JIRCAS) core project on Biological Nitrification Inhibition. The authors thank Dr. Shinya Iwasaki (JIRCAS) for helping to process some of the data in R.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised: Retrospective open access.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/bync/4.0/.
About this article
Cite this article
Sarr, P.S., Ando, Y., Nakamura, S. et al. Sorgoleone release from sorghum roots shapes the composition of nitrifying populations, total bacteria, and archaea and determines the level of nitrification. Biol Fertil Soils 56, 145–166 (2020). https://doi.org/10.1007/s00374-019-01405-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-019-01405-3