Abstract
Objectives
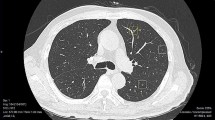
To compare the performance of radiologists in characterizing and diagnosing pulmonary nodules/masses with and without deep learning (DL)–based computer-aided diagnosis (CAD).
Methods
We studied a total of 101 nodules/masses detected on CT performed between January and March 2018 at Osaka University Hospital (malignancy: 55 cases). SYNAPSE SAI Viewer V1.4 was used to analyze the nodules/masses. In total, 15 independent radiologists were grouped (n = 5 each) according to their experience: L (< 3 years), M (3–5 years), and H (> 5 years). The likelihoods of 15 characteristics, such as cavitation and calcification, and the diagnosis (malignancy) were evaluated by each radiologist with and without CAD, and the assessment time was recorded. The AUCs compared with the reference standard set by two board-certified chest radiologists were analyzed following the multi-reader multi-case method. Furthermore, interobserver agreement was compared using intraclass correlation coefficients (ICCs).
Results
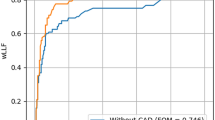
The AUCs for ill-defined boundary, irregular margin, irregular shape, calcification, pleural contact, and malignancy in all 15 radiologists, irregular margin and irregular shape in L and ill-defined boundary and irregular margin in M improved significantly (p < 0.05); no significant improvements were found in H. L showed the greatest increase in the AUC for malignancy (not significant). The ICCs improved in all groups and for nearly all items. The median assessment time was not prolonged by CAD.
Conclusions
DL-based CAD helps radiologists, particularly those with < 5 years of experience, to accurately characterize and diagnose pulmonary nodules/masses, and improves the reproducibility of findings among radiologists.
Key Points
• Deep learning–based computer-aided diagnosis improves the accuracy of characterizing nodules/masses and diagnosing malignancy, particularly by radiologists with < 5 years of experience.
• Computer-aided diagnosis increases not only the accuracy but also the reproducibility of the findings across radiologists.





Similar content being viewed by others
Abbreviations
- AI:
-
Artificial intelligence
- AUC:
-
Area under the receiver operating characteristic curve
- CAD:
-
Computer-aided diagnosis
- CT:
-
Computed tomography
- DL:
-
Deep learning
- ICC:
-
Intraclass correlation coefficients
- ROC:
-
Receiver operating characteristic
References
Siegel RL, Miller KD, Fuchs HE, Jemal A (2021) Cancer statistics, 2021. CA Cancer J Clin 71:7–33
National Lung Screening Trial Research Team, Aberle DR, Adams AM et al (2011) Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 365:395–409
Gould MK, Tang T, Liu IL et al (2015) Recent trends in the identification of incidental pulmonary nodules. Am J Respir Crit Care Med 192:1208–1214
Kuriyama K, Tateishi R, Doi O et al (1991) Prevalence of air bronchograms in small peripheral carcinomas of the lung on thin-section CT: comparison with benign tumors. AJR Am J Roentgenol 156:921–924
Li F, Sone S, Abe H, Macmahon H, Doi K (2004) Malignant versus benign nodules at CT screening for lung cancer: comparison of thin-section CT findings. Radiology 233:793–798
Ridge CA, Yildirim A, Boiselle PM et al (2016) Differentiating between subsolid and solid pulmonary nodules at CT: inter- and intraobserver agreement between experienced thoracic radiologists. Radiology 278(3):888–896
Lin H, Huang C, Wang W, Luo J, Yang X, Liu Y (2017) Measuring interobserver disagreement in rating diagnostic characteristics of pulmonary nodule using the lung imaging database consortium and image database resource initiative. Acad Radiol 24:401–410
Wataya T, Nakanishi K, Suzuki Y, Kido S, Tomiyama N (2020) Introduction to deep learning: minimum essence required to launch a research. Jpn J Radiol 38:907–921
White CS, Pugatch R, Koonce T, Rust SW, Dharaiya E (2008) Lung nodule CAD software as a Second Reader: a multicenter study. Acad Radiol 15:326–333
Higaki T, Nakamura Y, Zhou J et al (2020) Deep learning reconstruction at CT: phantom study of the image characteristics. Acad Radiol 27:82–87
Rudie JD, Gleason T, Barkovich MJ et al (2022) Clinical assessment of deep learning–based super-resolution for 3D volumetric brain MRI. Rad Artif Intell 4. https://doi.org/10.1148/ryai.210059
Hata A, Yanagawa M, Yamagata K et al (2021) Deep learning algorithm for detection of aortic dissection on non-contrast-enhanced CT. Eur Radiol 31:1151–1159
Peters AA, Huber AT, Obmann VC, Heverhagen JT, Christe A, Ebner L (2022) Diagnostic validation of a deep learning nodule detection algorithm in low-dose chest CT: determination of optimized dose thresholds in a virtual screening scenario. Eur Radiol. https://doi.org/10.1007/s00330-021-08511-7
Jiang W, Ganhua Z, Wang S, Wu X, Xu C (2022) Application of deep learning in lung cancer imaging diagnosis. J Healthc Eng 2022:6107940. https://doi.org/10.1155/2022/6107940
Ueda D, Yamamoto A (2021) Shimazaki A, et al Artificial intelligence-supported lung cancer detection by multi-institutional readers with multi-vendor chest radiographs: a retrospective clinical validation study. BMC Cancer 21:1120
Park HJ, Kim SM, La Yun B et al (2019) A computer-aided diagnosis system using artificial intelligence for the diagnosis and characterization of breast masses on ultrasound: added value for the inexperienced breast radiologist. Medicine (Baltimore) 98:e14146
Awai K, Murao K, Ozawa A et al (2006) Pulmonary nodules: estimation of malignancy at thin-section helical CT—effect of computer-aided diagnosis on performance of radiologists. Radiology 239:276–284
Yanagawa M, Niioka H, Kusumoto M et al (2021) Diagnostic performance for pulmonary adenocarcinoma on CT: comparison of radiologists with and without three-dimensional convolutional neural network. Eur Radiol 31:1978–1986
Hansell DM, Bankier AA, MacMahon H, McLoud TC, Müller NL, Remy J (2008) Fleischner Society: glossary of terms for thoracic imaging. Radiology 246:697–722
Muller NL, Silva CIS (2008) Imaging of the chest, vol 1. Saunders/Elsevier, Philadelphia
Xu Y, Ma L, Sun H et al (2020) CT-guided microcoil localization for pulmonary nodules before VATS: a retrospective evaluation of risk factors for pleural marking failure. Eur Radiol 30:5674–5683
Snoeckx A, Reyntiens P, Desbuquoit D et al (2018) Evaluation of the solitary pulmonary nodule: size matters, but do not ignore the power of morphology. Insights Imaging 9:73–86
Smith BJ, Hillis SL (2020) Multi-reader multi-case analysis of variance software for diagnostic performance comparison of imaging modalities. Proc SPIE Int Soc Opt Eng 11316. https://doi.org/10.1117/12.2549075
Anvari A, Halpern EF, Samir AE (2015) Statistics 101 for radiologists. Radiographics 35:1789–1801
Vallat R (2018) Pingouin: statistics in Python. J Open Source Softw 3:1026
Virtanen P, Gommers R, Oliphant TE et al (2020) SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 17:261–272
Eriguchi T, Takeda A, Tsurugai Y et al (2019) Pleural contact decreases survival in clinical T1N0M0 lung cancer patients undergoing SBRT. Radiother Oncol 134:191–198
Kim H-J, Cho JY, Lee YJ et al (2019) Clinical significance of pleural attachment and indentation of subsolid nodule lung cancer. Cancer Res Treat 51:1540–1548
Yagi T, Yamazaki M, Ohashi R et al (2018) HRCT texture analysis for pure or part-solid ground-glass nodules: distinguishability of adenocarcinoma in situ or minimally invasive adenocarcinoma from invasive adenocarcinoma. Jpn J Radiol 36:113–121
Kozuka T, Matsukubo Y, Kadoba T et al (2020) Efficiency of a computer-aided diagnosis (CAD) system with deep learning in detection of pulmonary nodules on 1-mm-thick images of computed tomography. Jpn J Radiol 38:1052–1061
Jin Z, Zhu Y, Zhang S et al (2020) Ultrasound computer-aided diagnosis (CAD) based on the Thyroid Imaging Reporting and Data System (TI-RADS) to distinguish benign from malignant thyroid nodules and the diagnostic performance of radiologists with different diagnostic experience. Med Sci Monit 26:e918452–e918452-11
Awai K, Murao K, Ozawa A et al (2004) Pulmonary nodules at chest CT: effect of computer-aided diagnosis on radiologists’ detection performance. Radiology 230:347–352
Song YS, Park CM, Park SJ, Lee SM, Jeon YK, Goo JM (2014) Volume and mass doubling times of persistent pulmonary subsolid nodules detected in patients without known malignancy. Radiology 273:276–284
Oda S, Awai K, Murao K et al (2011) Volume-doubling time of pulmonary nodules with ground-glass opacity at multidetector CT: assessment with computer-aided three-dimensional volumetry. Acad Radiol 18:63–69
Acknowledgements
This study was supported by FUJIFILM Corporation. The authors have been provided with the experimental picture archiving and communication systems and CAD.
Osaka University Reading Team is composed of the following radiologists. (Listed in alphabetical order)
Azusa Miura1, Hideyuki Fukui1, Kei Fujiwara2, Kengo Kiso1, Koki Kaketaka3, Masahiro Fujiwara1, Masataka Nakai2, Ryutaro Hosomi4, Shuhei Doi1, Takahisa Sakisuka1, Takashi Ota1, Tomo Miyata1, Toru Honda1, Yukihisa Sato5 and Yu Masuda6
1 Department of Radiology, Osaka University Graduate School of Medicine
2 Department of Radiation Oncology, Osaka University Graduate School of Medicine
3 Department of Diagnostic Radiology, Hyogo Prefectural Nishinomiya Hospital
4 Department of Diagnostic and Interventional Radiology, Kakogawa Municipal Central Hospital
5 Department of Radiology, Suita Municipal Hospital
6 Department of Diagnostic Imaging, Osaka General Medical Center
Funding
This study was supported by FUJIFILM Corporation. The authors have been provided with the experimental picture archiving and communication systems and CAD.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Shoji Kido, MD, PhD (Department of Artificial Intelligence Diagnostic Radiology, Osaka University Graduate School of Medicine).
Conflict of interest
The authors of this manuscript declare relationships with the following company: FUJIFILM Corporation.
Statistics and biometry
One of the authors has significant statistical expertise: Tomoharu Sato, PhD (Department of Biostatistics & Data Science, Osaka University Graduate School of Medicine).
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Approval for this study was obtained from the internal Ethics Review Board of Osaka University Hospital (Suita, Japan).
Methodology
• retrospective
• cross-sectional study
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wataya, T., Yanagawa, M., Tsubamoto, M. et al. Radiologists with and without deep learning–based computer-aided diagnosis: comparison of performance and interobserver agreement for characterizing and diagnosing pulmonary nodules/masses. Eur Radiol 33, 348–359 (2023). https://doi.org/10.1007/s00330-022-08948-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-022-08948-4