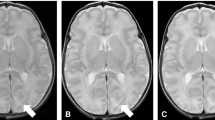
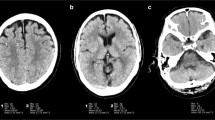
Abstract
Objectives
This study aimed to accelerate the 3D magnetization–prepared rapid gradient-echo (MPRAGE) sequence for brain imaging through the deep neural network (DNN).
Methods
This retrospective study used the k-space data of 240 scans (160 for the training set, mean ± standard deviation age, 93 ± 80 months, 94 males; 80 for the test set, 106 ± 83 months, 44 males) of conventional MPRAGE (C-MPRAGE) and 102 scans (77 ± 74 months, 52 males) of both C-MPRAGE and accelerated MPRAGE. All scans were acquired with 3T scanners. DNN was developed with simulated-acceleration data generated by under-sampling. Quantitative error metrics were compared between images reconstructed with DNN, GRAPPA, and E-SPIRIT using the paired t-test. Qualitative image quality was compared between C-MPRAGE and accelerated MPRAGE reconstructed with DNN (DNN-MPRAGE) by two readers. Lesions were segmented and the agreement between C-MPRAGE and DNN-MPRAGE was assessed using linear regression.
Results
Accelerated MPRAGE reduced scan times by 38% compared to C-MPRAGE (142 s vs. 320 s). For quantitative error metrics, DNN showed better performance than GRAPPA and E-SPIRIT (p < 0.001). For qualitative evaluation, overall image quality of DNN-MPRAGE was comparable (p > 0.999) or better (p = 0.025) than C-MPRAGE, depending on the reader. Pixelation was reduced in DNN-MPRAGE (p < 0.001). Other qualitative parameters were comparable (p > 0.05). Lesions in C-MPRAGE and DNN-MPRAGE showed good agreement for the dice similarity coefficient (= 0.68) and linear regression (R2 = 0.97; p < 0.001).
Conclusions
DNN-MPRAGE reduced acquisition time by 38% and revealed comparable image quality to C-MPRAGE.
Key Points
• DNN-MPRAGE reduced acquisition times by 38%.
• DNN-MPRAGE outperformed conventional reconstruction on accelerated scans (SSIM of DNN-MPRAGE = 0.96, GRAPPA = 0.43, E-SPIRIT = 0.88; p < 0.001).
• Compared to C-MPRAGE scans, DNN-MPRAGE showed improved mean scores for overall image quality (2.46 vs. 2.52; p < 0.001) or comparable perceived SNR (2.56 vs. 2.58; p = 0.08).






Similar content being viewed by others
Abbreviations
- 2D:
-
Two-dimensional
- 3D:
-
Three-dimensional
- ACS:
-
Autocalibrating signal
- C-MPRAGE:
-
Conventional image reconstruction of magnetization-prepared rapid gradient-echo images from conventional scans
- CNR:
-
Contrast-to-noise ratio
- CS:
-
Compressed sensing
- DNN:
-
Deep neural network
- DNN-MPRAGE:
-
Deep neural network–based reconstruction of magnetization-prepared rapid gradient-echo images from accelerated scans
- DSC:
-
Dice similarity coefficient
- E-SPIRIT:
-
An eigenvalue approach to iterative self-consistent parallel imaging reconstruction
- GRAPPA:
-
Generalized autocalibrating partially parallel acquisitions
- GW:
-
Gray-white matter
- MPRAGE:
-
Magnetization-prepared rapid gradient-echo
- MRI:
-
Magnetic resonance imaging
- NRMSE:
-
Normalized mean square errors
- PSNR:
-
Peak signal-to-noise ratio
- R :
-
Acceleration (or reduction) factor
- RSS:
-
Root sum of squares
- SENSE:
-
Sensitivity encoding
- SNR:
-
Signal-to-noise ratio
- SSIM:
-
Structural similarity index
- WAVE-CAIPI:
-
Wave-controlled aliasing in parallel imaging
References
Casey BJ, Giedd JN, Thomas KM (2000) Structural and functional brain development and its relation to cognitive development. Biol Psychol 54:241–257. https://doi.org/10.1016/s0301-0511(00)00058-2
Giedd JN, Rapoport JL (2010) Structural MRI of pediatric brain development: what have we learned and where are we going? Neuron 67:728–734. https://doi.org/10.1016/j.neuron.2010.08.040
Kim HG, Moon W-J, Han J, Choi JW (2017) Quantification of myelin in children using multiparametric quantitative MRI: a pilot study. Neuroradiology 59:1043–1051. https://doi.org/10.1007/s00234-017-1889-9
Kim HG, Choi JW, Han M et al (2020) Texture analysis of deep medullary veins on susceptibility-weighted imaging in infants: evaluating developmental and ischemic changes. Eur Radiol 30:2594–2603. https://doi.org/10.1007/s00330-019-06618-6
Mugler JP, Brookeman JR (1990) Three-dimensional magnetization-prepared rapid gradient-echo imaging (3D MP RAGE). Magn Reson Med 15:152–157. https://doi.org/10.1002/mrm.1910150117
Brant-Zawadzki M, Gillan GD, Nitz WR (1992) MP RAGE: a three-dimensional, T1-weighted, gradient-echo sequence--initial experience in the brain. Radiology 182:769–775. https://doi.org/10.1148/radiology.182.3.1535892
Mugler JP, Brookeman JR (1991) Rapid three-dimensional T1-weighted MR imaging with the MP-RAGE sequence. J Magn Reson Imaging 1:561–567. https://doi.org/10.1002/jmri.1880010509
Blumenthal JD, Zijdenbos A, Molloy E, Giedd JN (2002) Motion artifact in magnetic resonance imaging: implications for automated analysis. Neuroimage 16:89–92. https://doi.org/10.1006/nimg.2002.1076
Slovis TL (2011) Sedation and anesthesia issues in pediatric imaging. Pediatr Radiol 41:514. https://doi.org/10.1007/s00247-011-2115-2
Griswold MA, Jakob PM, Heidemann RM et al (2002) Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med 47:1202–1210. https://doi.org/10.1002/mrm.10171
Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P (1999) SENSE: sensitivity encoding for fast MRI. Magn Reson Med 42:952–962
Uecker M, Lai P, Murphy MJ et al (2014) ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: Where SENSE meets GRAPPA. Magn Reson Med 71:990–1001. https://doi.org/10.1002/mrm.24751
Nana R, Zhao T, Heberlein K et al (2008) Cross-validation-based kernel support selection for improved GRAPPA reconstruction. Magn Reson Med 59:819–825. https://doi.org/10.1002/mrm.21535
Lustig M, Pauly JM (2010) SPIRiT: iterative self-consistent parallel imaging reconstruction from arbitrary k-space. Magn Reson Med 64:457–471. https://doi.org/10.1002/mrm.22428
Crémillieux Y, Briguet A, Deguin A (1994) Projection-reconstruction methods: fast imaging sequences and data processing. Magn Reson Med 32:23–32. https://doi.org/10.1002/mrm.1910320105
Bilgic B, Gagoski BA, Cauley SF et al (2014) Wave-CAIPI for highly accelerated 3D imaging. Magn Reson Med 73:2152–2162. https://doi.org/10.1002/mrm.25347
Lustig M, Donoho DL, Santos JM, Pauly JM (2008) Compressed sensing MRI. IEEE Signal Process Mag 25:72–82. https://doi.org/10.1109/msp.2007.914728
Cheng JY, Mardani M, Alley MT et al (2018) DeepSPIRiT: generalized parallel imaging using deep convolutional neural networks. In: Proceedings of the 26th Annual Meeting of ISMRM. Paris, France, p 0570. https://cds.ismrm.org/protected/18MPresentations/abstracts/0570.html
Sriram A, Zbontar J, Murrell T, et al (2020) End-to-end variational networks for accelerated MRI reconstruction. arXiv Prepr arXiv:200406688
Sriram A, Zbontar J, Murrell T, et al (2020) GrappaNet: combining parallel imaging with deep learning for multi-coil MRI reconstruction. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 14315–14322. https://openaccess.thecvf.com/content_CVPR_2020/html/Sriram_GrappaNet_Combining_Parallel_Imaging_With_Deep_Learning_for_Multi-Coil_MRI_CVPR_2020_paper.html
Eo T, Jun Y, Kim T et al (2018) KIKI-net: cross-domain convolutional neural networks for reconstructing undersampled magnetic resonance images. Magn Reson Med 80:2188–2201. https://doi.org/10.1002/mrm.27201
Hammernik K, Klatzer T, Kobler E et al (2017) Learning a variational network for reconstruction of accelerated MRI data. Magn Reson Med 79:3055–3071. https://doi.org/10.1002/mrm.26977
Aggarwal HK, Mani MP, Jacob M (2019) MoDL: model-based deep learning architecture for inverse problems. IEEE Trans Med Imaging 38:394–405. https://doi.org/10.1109/tmi.2018.2865356
Muckley MJ, Riemenschneider B, Radmanesh A et al (2021) Results of the 2020 fastMRI challenge for machine learning MR image reconstruction. IEEE Trans Med Imaging 40:2306–2317. https://doi.org/10.1109/tmi.2021.3075856
Akçakaya M, Moeller S, Weingärtner S, Uğurbil K (2019) Scan-specific robust artificial-neural-networks for k-space interpolation (RAKI) reconstruction: database-free deep learning for fast imaging. Magn Reson Med 81:439–453. https://doi.org/10.1002/mrm.27420
Girshick R, Donahue J, Darrell T, Malik J (2014) Rich feature hierarchies for accurate object detection and semantic segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 580–587. https://openaccess.thecvf.com/content_cvpr_2014/html/Girshick_Rich_Feature_Hierarchies_2014_CVPR_paper.html
He L, Wang J, Lu Z-L et al (2018) Optimization of magnetization-prepared rapid gradient echo (MP-RAGE) sequence for neonatal brain MRI. Pediatr Radiol 48:1139–1151. https://doi.org/10.1007/s00247-018-4140-x
Wang Z, Bovik AC, Sheikh HR, Simoncelli EP (2004) Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process 13:600–612. https://doi.org/10.1109/tip.2003.819861
Acharya UR, Anand D, Bhat PS, Niranjan UC (2001) Compact storage of medical images with patient information. IEEE Trans Inf Technol Biomed 5:320. https://doi.org/10.1109/4233.966107
Almohammad A, Ghinea G (2010) Stego image quality and the reliability of PSNR. In: 2010 2nd International Conference on Image Processing Theory, Tools and Applications. pp 215–220. https://doi.org/10.1109/IPTA.2010.5586786
Fenster A, Chiu B (2005) Evaluation of segmentation algorithms for medical imaging. In: Conf Proc IEEE Eng Med Biol Soc. Shanghai, pp 7186–7189. https://doi.org/10.1109/IEMBS.2005.1616166
Kozak BM, Jaimes C, Kirsch J, Gee MS (2020) MRI techniques to decrease imaging times in children. Radiographics 40:485–502. https://doi.org/10.1148/rg.2020190112
Ji S, Jeong J, Oh S-H et al (2021) Quad-contrast imaging: simultaneous acquisition of four contrast-weighted images (PD-weighted, T2-weighted, PD-FLAIR and T2-FLAIR images) with synthetic T1-weighted image, T1-and T2-maps. IEEE Trans Med Imaging 40:3617–3626. https://doi.org/10.1109/tmi.2021.3093617
Kim KH, Choi SH, Park S-H (2018) Improving arterial spin labeling by using deep learning. Radiology 287:658–666. https://doi.org/10.1148/radiol.2017171154
Fujita S, Hagiwara A, Otsuka Y et al (2020) Deep learning approach for generating MRA images from 3D quantitative synthetic MRI without additional scans. Invest Radiol 55:249–256. https://doi.org/10.1097/rli.0000000000000628
Williams L-A, DeVito TJ, Winter JD et al (2007) Optimization of 3D MP-RAGE for neonatal brain imaging at 3.0 T. Magn Reson Imaging 25:1162–1170. https://doi.org/10.1016/j.mri.2007.01.119
Kaye EA, Aherne EA, Duzgol C et al (2020) Accelerating prostate diffusion-weighted MRI using a guided denoising convolutional neural network: retrospective feasibility study. Radiology Artif Intell 2:e200007. https://doi.org/10.1148/ryai.2020200007
Koonjoo N, Zhu B, Bagnall GC et al (2021) Boosting the signal-to-noise of low-field MRI with deep learning image reconstruction. Sci Rep 11:8248. https://doi.org/10.1038/s41598-021-87482-7
Montejo C, Laredo C, Llull L et al (2021) Synthetic MRI in subarachnoid haemorrhage. Clin Radiol 76:785.e17–785.e23. https://doi.org/10.1016/j.crad.2021.05.021
Fujita S, Yokoyama K, Hagiwara A et al (2021) 3D quantitative synthetic MRI in the evaluation of multiple sclerosis lesions. AJNR Am J Neuroradiol 42:471–478. https://doi.org/10.3174/ajnr.a6930
Kellman P, McVeigh ER (2005) Image reconstruction in SNR units: a general method for SNR measurement†. Magn Reson Med 54:1439–1447. https://doi.org/10.1002/mrm.20713
Tukey JW (1967) An introduction to the calculations of numerical spectrum analysis. Spectra Analysis of Time Series 25–46. https://ci.nii.ac.jp/naid/10011111666/#cit
Keil B, Alagappan V, Mareyam A et al (2011) Size-optimized 32-channel brain arrays for 3 T pediatric imaging. Magn Reson Med 66:1777–1787. https://doi.org/10.1002/mrm.22961
Kim M, Kim HS, Kim HJ et al (2021) Thin-slice pituitary MRI with deep learning–based reconstruction: diagnostic performance in a postoperative setting. Radiology 298:114–122. https://doi.org/10.1148/radiol.2020200723
Herrmann J, Gassenmaier S, Nickel D et al (2020) Diagnostic confidence and feasibility of a deep learning accelerated HASTE sequence of the abdomen in a single breath-hold. Invest Radiol 56:313–319. https://doi.org/10.1097/rli.0000000000000743
Kidoh M, Shinoda K, Kitajima M et al (2020) Deep learning based noise reduction for brain MR imaging: tests on phantoms and healthy volunteers. Magn Reson Med Sci 19:195–206. https://doi.org/10.2463/mrms.mp.2019-0018
Gassenmaier S, Afat S, Nickel MD et al (2021) Accelerated T2-weighted TSE imaging of the prostate using deep learning image reconstruction: a prospective comparison with standard T2-weighted TSE imaging. Cancers 13:3593. https://doi.org/10.3390/cancers13143593
Ueda T, Ohno Y, Yamamoto K et al (2021) Compressed sensing and deep learning reconstruction for women’s pelvic MRI denoising: utility for improving image quality and examination time in routine clinical practice. Eur J Radiol 134:109430. https://doi.org/10.1016/j.ejrad.2020.109430
Funding
This work was supported by the Korea Medical Device Development Fund grant funded by the Korean government (the Ministry of Science and ICT, the Ministry of Trade, Industry and Energy, the Ministry of Health & Welfare, the Ministry of Food and Drug Safety) (Project Number: KMDF_PR_20200901_0062, 9991006735) and the National Research Foundation of Korea (NRF) grant funded by the Korean government (Project Number: NRF-2021R1A2C1007831).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Hyun Gi Kim.
Conflict of interest
Three of the authors of this manuscript declare relationships with AIRS Medical Inc.
Woojin Jung, Jingyu Ko, and Geunu Jeong are employees of AIRS Medical.
The other authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and Biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethics approval
Institutional Review Board approval was obtained.
Methodology
• Retrospective
• Cross-sectional study
• Performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 3023 kb)
Rights and permissions
About this article
Cite this article
Jung, W., Kim, J., Ko, J. et al. Highly accelerated 3D MPRAGE using deep neural network–based reconstruction for brain imaging in children and young adults. Eur Radiol 32, 5468–5479 (2022). https://doi.org/10.1007/s00330-022-08687-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-022-08687-6