Abstract
Objectives
To investigate machine learning classifiers and interpretable models using chest CT for detection of COVID-19 and differentiation from other pneumonias, interstitial lung disease (ILD) and normal CTs.
Methods
Our retrospective multi-institutional study obtained 2446 chest CTs from 16 institutions (including 1161 COVID-19 patients). Training/validation/testing cohorts included 1011/50/100 COVID-19, 388/16/33 ILD, 189/16/33 other pneumonias, and 559/17/34 normal (no pathologies) CTs. A metric-based approach for the classification of COVID-19 used interpretable features, relying on logistic regression and random forests. A deep learning–based classifier differentiated COVID-19 via 3D features extracted directly from CT attenuation and probability distribution of airspace opacities.
Results
Most discriminative features of COVID-19 are the percentage of airspace opacity and peripheral and basal predominant opacities, concordant with the typical characterization of COVID-19 in the literature. Unsupervised hierarchical clustering compares feature distribution across COVID-19 and control cohorts. The metrics-based classifier achieved AUC = 0.83, sensitivity = 0.74, and specificity = 0.79 versus respectively 0.93, 0.90, and 0.83 for the DL-based classifier. Most of ambiguity comes from non-COVID-19 pneumonia with manifestations that overlap with COVID-19, as well as mild COVID-19 cases. Non-COVID-19 classification performance is 91% for ILD, 64% for other pneumonias, and 94% for no pathologies, which demonstrates the robustness of our method against different compositions of control groups.
Conclusions
Our new method accurately discriminates COVID-19 from other types of pneumonia, ILD, and CTs with no pathologies, using quantitative imaging features derived from chest CT, while balancing interpretability of results and classification performance and, therefore, may be useful to facilitate diagnosis of COVID-19.
Key Points
• Unsupervised clustering reveals the key tomographic features including percent airspace opacity and peripheral and basal opacities most typical of COVID-19 relative to control groups.
• COVID-19-positive CTs were compared with COVID-19-negative chest CTs (including a balanced distribution of non-COVID-19 pneumonia, ILD, and no pathologies). Classification accuracies for COVID-19, pneumonia, ILD, and CT scans with no pathologies are respectively 90%, 64%, 91%, and 94%.
• Our deep learning (DL)–based classification method demonstrates an AUC of 0.93 (sensitivity 90%, specificity 83%). Machine learning methods applied to quantitative chest CT metrics can therefore improve diagnostic accuracy in suspected COVID-19, particularly in resource-constrained environments.
Similar content being viewed by others
Introduction
Coronavirus disease 2019 or COVID-19 has caused a global pandemic associated with an immense human toll and healthcare burden across the world [1]. COVID-19 can manifest as pneumonia, which may lead to acute hypoxemic respiratory failure, the main reason for hospitalization and mortality. A Fleischner Society statement supports the use of lung imaging for differential diagnosis and management of patients with moderate to severe clinical symptoms, especially in resource-constrained environments [2]. The most typical pulmonary CT imaging features of COVID-19 are multifocal (often bilateral and peripheral predominant) airspace opacities, comprised of ground glass opacities and/or consolidation, which may be associated with interlobular and intralobular septal thickening [3]. A study comparing COVID-19 and other types of viral pneumonia demonstrated that distinguishing features more typical of COVID-19 are the predominance of ground glass opacities, peripheral distribution, and perivascular thickening [4]. A consensus statement on COVID-19 reporting by the Radiological Society of North America indicates the typical appearance of COVID-19 as peripheral and bilateral distribution of ground glass opacities with or without consolidation, and possibly with the “reverse halo” sign [5]. Confirmatory diagnosis of COVID-19 requires identification of the virus on nasopharyngeal swabs via RT-PCR (reverse transcription polymerase chain reaction), a highly specific test (>99%) but with lower sensitivity (50–80%) [6, 7]. Given the imperfect sensitivity of RT-PCR and potential resource constraints, chest CT imaging has an evolving role in diagnosis of COVID-19 and possibly prognostic value.
Recently, several groups have shown that COVID-19 can be identified on CT with variable accuracy. For example, chest CTs in patients who were positive for COVID-19 (RT-PCR) could be distinguished from chest CTs in patients that tested negative with an AUC of 0.92 using machine learning [8]. While this classification is potentially valuable, it is limited by a lack of details on the types and distribution of findings on negative (control) cases. It is important to distinguish COVID-19-related pulmonary disease not only from subjects with no pathologies CTs but also from other types of lung diseases unrelated to COVID-19, including other infections, malignancy, interstitial lung disease (ILD), and chronic obstructive pulmonary disease (COPD). This is especially important as COVID-19 can manifest similarly clinically to other respiratory infections such as influenza, which can lead to confusion in triage and diagnosis. Bai et al showed that an artificial intelligence system can assist radiologists to distinguish COVID-19 from other types of pneumonia, with a diagnostic sensitivity of 88% and specificity of 90% [9]. The two cohorts (COVID-19 and other pneumonia) compared in this study are from two different countries, limiting the generalizability of their model. Other studies showing promising results in classification do not provide a detailed description of imaging cohorts acquisition protocols or data sources [10, 11]. This information is important since different institutions will have diverse CT acquisition protocols and clinical indications for CT usage, which can affect the performance of machine learning algorithms.
Our goals were to compute CT-derived quantitative imaging metrics corresponding to the typical presentation of COVID-19 and evaluate the discriminative power of these metrics for the diagnosis of COVID-19, when compared to different compositions of control groups; perform unsupervised clustering of interpretable features to visualize how COVID-19 patients differ from controls; compare the performance of metrics-based classifiers to a deep learning–based model. Our large training and test datasets contained chest CTs in patients confirmed with COVID-19 and negative controls from multiple institutions in North America and Europe, making this one of the first large studies to demonstrate the value of machine learning for differentiation of COVID-19 and non-COVID-19 utilizing data from multiple centers, increasing generalizability and applicability.
Methods
Patient selection and imaging data
This retrospective study utilized data acquired from 16 different centers in North America and Europe, after anonymization, ethical review and approval at each institution. Our dataset consists of chest CTs of 1226 patients positive for COVID-19, and 1287 chest CTs of patients without COVID-19, including other types of pneumonia (n = 240), ILD (n = 437), and without any pathologies on chest CT (n = 610). The flowchart for patient selection criteria is shown in Fig. 1. All CTs in the COVID-19 cohort from North America have been confirmed by RT-PCR. The COVID-19 cohort from Europe has been either confirmed by RT-PCR or diagnosed based on clinical symptoms, epidemiological exposure, and radiological assessment. The pneumonia cohort consists of cases of patients with non-COVID-19 viral or bacterial pneumonias, organizing pneumonia, and aspiration pneumonia. The ILD cohort consists of patients with various types of ILD such as usual interstitial pneumonia, nonspecific interstitial pneumonia, and other unclassifiable interstitial diseases with or without fibrotic features, which exhibit ground glass opacities, reticulation, honeycombing, traction bronchiectasis, and consolidation to different degrees. Sixty-four COVID-19 cases were excluded due to no opacities on chest CT, 84 COVID-19 cases were excluded due to had minimal opacities (PO < 1%), one COVID-19 case was excluded due to incomplete inclusion of the lungs in the field-of-view, and two pneumonia controls were excluded due to incorrect DICOM parameters and imaging artifacts.
The dataset was divided into training (2147), validation (99), and test (200) sets (Table 1). Model training and selection were performed based on training and validation sets. The final performance of selected models is reported on the test dataset (Table S1 provides a detailed breakdown of demographic and scanning information for each cohort).
Metrics of airspace disease severity
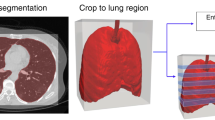
We computed thirty-two metrics of severity based on abnormalities known to be associated with COVID-19, as well as lung and lobar segmentation, using a previously developed Deep Image-to-Image Network trained on a large cohort of healthy and abnormal cases for lung segmentation [12]. Next, we used a DenseUnet to identify abnormalities such as GGO and consolidations on COVID-19 as well as control groups [12]. Based on these segmentations, we computed severity metrics to summarize the spatial distribution and extent of airspace disease in both lungs. A complete and detailed description of the thirty-two metrics is provided in Table S3. Our algorithm is fully automated and requires no manual input (Fig. 2).
Overview of the deep learning–based COVID-19 classifier. Preprocessing consists of lung segmentation and opacities probability distribution computation [12] followed by a 3D deep neural network trained to distinguish between the COVID-19 class and non-COVID-19 class
Metric-based analysis
Unsupervised feature selection and clustering
Recursive feature elimination was used to select the metrics of severity most discriminative between COVID-19 and non-COVID-19 classes. The k best features were selected based on an internal validation split. Based on the selected metrics, an unsupervised hierarchical cluster analysis was performed to identify clusters of images that have similar features. The pairwise Euclidean distance between two metrics was used to compute a distance matrix, with average linkage method used for hierarchical clustering [13], visualized as a heatmap (Fig. 3).
Heat map of hierarchical clustering. This illustrates the unsupervised hierarchical clustering of the seven metrics along with cohort membership (COVID-19, other pneumonia, ILD, and no pathologies) from the entire training set of 1800 cases. The metric values are standardized and rescaled to a value between 0 and 1. a Training dataset; b Test dataset
Supervised COVID-19 classification
Two metrics-based classifiers were trained based on the thirty-two computed metrics. First, we trained a random forest classifier, M1, using k-selected features based on recursive feature elimination. Subsequently, we trained a second classifier using logistic regression (LR), after feature transformation based on gradient boosted trees (GBT) [14]. For training GBT, we used 2000 estimators with max depth = 3 and 3 features for each split. The boosting fraction 0.8 was used for fitting the individual trees. The logistic regression classifier, M2, was trained with L2 regularization (C = 0.2). The class weights were adjusted to class frequencies for the class imbalance between COVID-19 and non-COVID-19 classes.
Supervised deep learning–based COVID-19 classification
A deep learning–based 3D neural network model, M3, was trained to separate the positive class (COVID-19) vs negative class (non-COVID-19). As input, we considered a two-channel 3D tensor, with the first channel containing directly the CT Hounsfield units within the lung segmentation masks and the second channel containing the probability map of a previously proposed opacity classifier [12]. The 3D network uses anisotropic 3D kernels to balance resolution and speed with deep dense blocks that gradually aggregate features down to a binary output. The network was trained end-to-end as a classification system using binary cross-entropy and uses probabilistic sampling of the training data to adjust for the imbalance in the training dataset labels. A separate validation dataset was used for final model selection before the performance was measured on the testing set. The input 3D tensor size is fixed (2 × 128 × 384 × 384) corresponding to the lung segmentation from the CT data rescaled to a 3×1×1 mm resolution. The first two blocks are anisotropic and consist of convolution (kernels 1 × 3 × 3)—batch normalization—LeakyReLU and Max-pooling (kernels 1 × 2 × 2, stride 1 × 2 × 2). The subsequent five blocks are isotropic with convolution (kernels 3 × 3 × 3)—batch normalization—LeakyReLU and Max-pooling (kernels 2 × 2 × 2, stride 2 × 2 × 2) followed by a final linear classifier with the input 144-dimensional. Figure 2 depicts our 3D DL classifier.
Comparison with models from the literature and with radiologist performance
We compared the models in this work to those published by Li et al [10] and the model proposed by Harmon et al [15]. Li et al [10] investigated a deep learning method to distinguish COVID-19 from community-acquired pneumonia and healthy subjects using chest CT. Their proposed DL method is based on extracting 2D features on each CT slice followed by feature pooling across slices and a final linear classifier. There are two main differences between the DL method proposed in this article and the one proposed by Li et al [10]. First, our method is based on 3D deep learning, which better leverages the contiguity of imaging textures along the z-axis, and second, it uses as input the spatial distribution of opacities within the lung parenchyma, which focuses the classifier on the regions of abnormality. For the method of Li et al [10], we have re-trained the model on our training dataset while for Harmon et al [15], we have run their released model on our testing set.
We have investigated how our method compares against the CO-RADS scoring system [16]. CO-RADS is a categorical score that quantifies the suspicion of pulmonary involvement by COVID-19 and was shown to perform well in predicting disease severity of COVID-19 patients. To understand how the CO-RADS scoring system performs in our test set ground truth, we have asked a radiologist (with over 5 years of experience) to score the 200 test cases. The radiologist was provided only with chest CT scans and not provided with any additional information about the ground truth diagnosis or RT-PCR test.
Results
Six features were selected by recursive feature elimination between features and classes in the training dataset of 1011 COVID-19 cases and 1136 controls (other pneumonia, ILD, and no pathologies). The features are as follows:
-
1.
Percent of ground glass opacities
-
2.
Percent of opacity (PO) (consolidation and ground glass opacities = airspace disease)
-
3.
Percent of opacities in the periphery (see Appendix)
-
4.
Percent of opacities in the rind (see Appendix)
-
5.
Percent of opacities in the right lower lobe
-
6.
Percent of opacities in the left lower lobe
Figure 3 demonstrates the hierarchical clustering of these metrics, along with the ground truth diagnosis cohort membership (COVID-19, other pneumonia, ILD, and no pathologies CT) shown on the band on the left of heat map. The metric values are standardized and rescaled to a value between 0 and 1. In Fig. 3a, the clustering is performed on the entire training set of 1800 subjects. The probability of belonging to the COVID-19 class increases towards the bottom of the heat map, which corresponds to higher values of the metrics, i.e., more opacities (both GGO and consolidation), and more peripheral and basal distribution. The middle of the heatmap shows the ambiguous region, where there is an overlap of features from different disease cohorts. Figure 3b shows the same clustering in the test dataset for each of the disease cohorts. While there is a cluster of COVID-19 subjects that have characteristic features, there are also many which do not show all characteristics. Moreover, some cases of pneumonia and ILD overlap with the typical features of COVID-19.
The six selected features were used to train a random forest classifier (M1). The performance of this classifier on a test dataset has an AUC of 0.75 (95% CI: [0.69, 0.81]) as depicted in Fig. 4, which shows bootstrapped ROC and AUC values, along with their 95% confidence intervals, which were computed on 1000 samples with replacement. The sensitivity and specificity of this model are 0.86 and 0.60, respectively. The performance is improved by training a second classifier on all thirty-two metrics using a logistic regression model (M2). The metrics are first transformed to a higher-dimensional space using feature embedding with gradient boosted trees. On the test set, this model produces an AUC of 0.83 (95% CI: [0.78, 0.89]) with a sensitivity of 0.74 and a specificity of 0.79. While the performance improves, some of the interpretability is lost since the features are transformed to a higher dimension.
a Bootstrapped ROCs for discriminating COVID-19 from ILD, other pneumonia, and no pathology control by the models proposed in this study. The models M1, M2, and M3 and CO-RADS scoring [16] were evaluated with 100 COVID-19 positive, 33 ILD, 33 other pneumonia, and 34 healthy without pathologies on CTs. The 95% confidence intervals (shown as a band) are computed by bootstrapping over 1000 samples with replacement from the predicted scores. b Bootstrapped ROCs for our 3D DL classifier (M3), the model proposed by Li et al [10], and the model proposed by Harmon et al [15]. For the model proposed by Li et al, we trained and tested on our dataset using the code provided by the authors. The 95% confidence intervals (shown as a band) are computed by bootstrapping over 1000 samples with replacement from the predicted scores
Our deep learning–based classifier (M3) has the best performance with an AUC of 0.93 (95% CI: [0.90, 0.96]), improving the sensitivity and specificity of the system to 0.90 and 0.83, respectively. The improvement is mostly due to a reduction of false positives from the ILD and an increase of true positives in the COVID-19 class. The optimal operating point for all models was chosen as the point with the shortest distance from the top left corner on the ROC computed on the whole test dataset, without bootstrapping. The corresponding confusion matrices for the three models are shown in Table 2. Figure 5 illustrates examples of correctly labeled samples by the metrics-based classifier and the DL-based classifier, on typical CT images from COVID-19 patients. Figure S2 shows negative examples from ILD and non-COVID-19 pneumonia patients. Overlaid in red are the areas identified by the opacity classifier. Figure 6 illustrates examples of cases incorrectly labeled by both classifiers and Figure S3 shows cases that are incorrectly labeled by the metric-based classifier but correctly labeled by the DL classifier that uses additional texture features extracted directly from the images.
Examples of incorrectly classified samples by both methods: top-row COVID-19 (false negative), middle-row ILD (false positive), bottom-row other pneumonia (false positive). Red marks abnormalities associated with COVID-19 (top-row), associated with ILD (middle-row), or associated with other pneumonia (bottom-row), respectively
The performance of the CO-RADS scoring system is illustrated in Fig. 4 and Table 2. For the sake of our comparison, we consider that CO-RADS 4-5 is a positive prediction for COVID-19 and CO-RADS 1-3 as negative prediction. The performance of the CO-RADS scoring system is comparable to the metric-based classifiers.
We compared the models in this work to the models previously published by Li et al and Harmon et al on our test dataset. The model from Li et al achieved an AUC of 0.90 (95% CI: [0.86, 0.94]) and the model from Harmon et al achieved an AUC of 0.74 (95% CI: [0.67, 0.81]) on our testing set as shown in Figure S2. The optimal operating point, which was selected as the point closest to the top left corner of the ROC computed on the whole test dataset, without bootstrapping, produced a sensitivity of 0.86 and specificity of 0.80 for Li et al and a sensitivity of 0.64 and specificity of 0.78 for Harmon et al (Fig. 4b). The confusion matrix is shown in Table 3.
We have computed the potential impact of technical parameters of the acquisition on the sensitivity of the deep learning-based classifier M3. The results for the parameters where there is significant data support are illustrated in Table S2. The table illustrates good performance (90.2%, 93.8%) for COVID-19 detection for high-resolution acquisitions (slice thickness below 3 mm) while the performance degrades (72.7%) to lower resolution acquisitions (slice thickness above 3 mm). Soft and hard kernel types are evenly represented in the test data with slightly higher performance on soft kernels (92.0%) than on hard kernels (91.2%) which could be attributed to lower noise levels. Sensitivity across manufacturers is on average 92.75% (+7.25/−4.15) and illustrated in Table S2.
To understand the limitation of detecting COVID-19 on datasets without visible airspace opacities on chest CT, we have analyzed the performance of the deep learning–based classifier M3 on the 64 COVID-19 datasets that were excluded from the main data selection process. The M3 classifier predicted 33 cases as COVID-19 positive (51.6%) and 30 cases as COVID-19 negative (48.4%). This performance suggests that even if there are no visible opacities, there are other imaging features that can distinguish COVID-19 from other diseases.
We have assessed if patients in our cohort had a co-existence of disease patterns such as COVID-19 superimposed with ILD or emphysema. Upon review, we have identified only 2 COVID-19-positive cases with underlying ILD (Figure S4) and 1 COVID-19-positive case with underlying Emphysema. The first 2 are predicted as positive by all three models while the third is predicted positive only by the M3 model.
Discussion
We evaluated the ability of machine learning algorithms to distinguish between chest CTs in patients with COVID-19 and a diverse control cohort comprising of chest CTs demonstrating other pneumonias, ILD, and no pathology. We performed an analysis based on clinically interpretable severity metrics computed from automated segmentation of abnormal regions in a chest CT scan, as well as using a deep learning system. The six selected features by recursive elimination correspond to reported typical COVID-19 features: multifocal ground glass opacities and/or consolidation with basal and peripheral distribution [5, 16, 17]. Unsupervised clustering on selected severity metrics shows that while there are dominant characteristics that can be observed in COVID-19 such as ground glass opacities as well as peripheral and basal distribution, these are not observed in all cases of COVID-19. On the other hand, some ILD and other pneumonia patients can exhibit similar characteristics. We found that the performance of the system can be improved by mapping these metrics into a higher-dimensional space prior to training a classifier, as shown by model M2 in Fig. 3. The best classification performance, with sensitivity/specificity of 90%/83% is achieved by the deep learning system M3, which is essentially a high-dimensional, non-linear model. For reference, we have also included the performance of the manually rated CO-RADS scoring system by a radiologist on the same test set, which had sensitivity/specificity of 74%/71%. The reported performance of CO-RADS in identifying COVID-19 (positive RT-PCR) is variable in literature with Bellini et al reporting sensitivity/specificity of 71.0%/81.0% [18], Lieveld et al reporting sensitivity/specificity of 89.4%/87.2% [19], and Prokop et al reporting sensitivity/specificity of 90%/82.8% [16].
The deep learning method achieves reduced false-positive and false-negative rates relative to the metrics-based classifier suggesting that there might be other latent radiological manifestations of COVID-19 that distinguish it from ILDs or other types of pneumonia. It is worth investigating how to incorporate the common imaging features into our 3D DL classifier as prior information. The proposed AI-based method has been trained and tested on a database of 2446 CT datasets with 1161 COVID-19 patients and 1285 datasets coming from other categories. We also show how our method compares to the one published by Li et al [10] and found that our method achieves a higher AUC as well as sensitivity. Further details are provided in the supplementary section.
One limitation of this study is that our training set is biased toward COVID-19 and normal controls, potentially affecting discrimination of other lung pathologies. Another limitation is that the validation set size is relatively small, which might not capture the entire data distribution of clinical use cases for optimal model selection. Among the strengths of this study are the diversity of training and testing CTs used, acquired using a variety of CT scanners, from numerous institutions and regions, ensuring robust and generalizable results. We also included not only normal controls but also various types of lung pathology in the COVID-19-negative group. We have included an analysis of the system sensitivity to main acquisition parameters.
Our system provides clinical value in several aspects: it can be used for rapid triage of positive cases, particularly in resource-constrained environments where radiologic expertise or RT-PCR may not be immediately available; it could help radiologists to prioritize interpreting CTs in patients with COVID-19 by screening out lower probability cases. The output of our deep learning classifier is easily reproducible and replicable, mitigating inter-reader variability. While RT-PCR will remain the reference standard for confirmatory diagnosis of COVID-19, machine learning methods applied to quantitative CT can perform with high diagnostic accuracy, increasing the value of imaging in the diagnosis and management of this disease. In addition, the ability of the deep learning classifier to detect COVID-19 cases on datasets with no visible opacities on CT images suggests that there are additional image features beyond airspace disease that could be used to detect COVID-19 and should be investigated in the future. Furthermore, these algorithms could be integrated into a surveillance effort for COVID-19, even in unsuspected patients, in high-incidence regions, with automatic assessment for evidence of COVID-19 lung disease, allowing more rapid institution of isolation protocols. Finally, it could potentially be applied retrospectively to large numbers of chest CT exams from institutional PACS systems worldwide to uncover the dissemination of SARS-CoV-2 in communities prior to the implementation of widespread testing efforts.
In the future, we plan to deploy and validate the algorithm in a clinical setting and evaluate the clinical utility and diagnostic accuracy on prospective data, as well as to investigate the value to predict clinical severity and prognosis of COVID-19, as well as ancillary findings of COVID-19 such as acute pulmonary embolism, which is associated with severe COVID-19 [20, 21]. In addition, clinical decision models could be improved by training a classifier that incorporates other clinical data such as pulse oximetry and laboratory metrics, in addition to imaging features.
Abbreviations
- AUC:
-
Area under the curve
- CO-RADS:
-
COVID-19 Reporting and Data System
- COVID-19:
-
Coronavirus Disease 2019
- DICOM:
-
Digital Imaging and Communications in Medicine
- DL:
-
Deep learning
- GBT:
-
Gradient boosted trees
- GGO:
-
Ground glass opacity
- ILD:
-
Interstitial lung disease
- LR:
-
Logistic regression
- PACS:
-
Picture Archiving and Communication System
- PHO:
-
Percent of high opacity
- PO:
-
Percent of opacity
- RF:
-
Random forest
- ROC:
-
Receiver operating characteristic
- RT-PCR:
-
Reverse transcript polymerase chain reaction
- SARS-CoV-2:
-
Severe Acute Respiratory Syndrome Coronavirus 2
References
JHU. Coronavirus COVID-19 Global Cases by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU) [Internet]. 2020. Available from: https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6
Rubin GD, Ryerson CJ, Haramati LB et al (2020) The role of chest imaging in patient management during the COVID-19 pandemic: a multinational consensus statement from the Fleischner Society. Chest.
Bernheim A, Mei X, Huang M et al (2020) Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. 200463
Bai HX, Hsieh B, Xiong Z et al (2020) Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology. 200823
Simpson S, Kay FU, Abbara S et al (2020) Radiological Society of North America Expert Consensus Statement on Reporting Chest CT Findings Related to COVID-19. Endorsed by the Society of Thoracic Radiology, the American College of Radiology, and RSNA. Radiol Cardiothorac Imaging 2(2):e200152
Kanne JP, Little BP, Chung JH, Elicker BM, Ketai LH (2020) Essentials for radiologists on COVID-19: an update—radiology scientific expert panel. Radiology. 200527
Fang Y, Zhang H, Xie J et al (2020) Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 200432
Mei X, Lee H-C, Diao K et al (2020) Artificial intelligence-enabled rapid diagnosis of COVID-19 patients. medRxiv
Bai HX, Wang R, Xiong Z et al (2020) AI augmentation of radiologist performance in distinguishing COVID-19 from pneumonia of other etiology on chest CT. Radiology. 201491
Li L, Qin L, Xu Z et al (2020) Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 200905
Singh D, Kumar V, Kaur M (2020) Classification of COVID-19 patients from chest CT images using multi-objective differential evolution–based convolutional neural networks. Eur J Clin Microbiol Infect Dis:1–11
Chaganti S, Balachandran A, Chabin G et al (2020) Quantification of tomographic patterns associated with COVID-19 from chest CT. arXiv Prepr arXiv200401279
Müllner D (2011) Modern hierarchical, agglomerative clustering algorithms. arXiv Prepr arXiv11092378
Friedman JH (2001) Greedy function approximation: a gradient boosting machine. Ann Stat:1189–1232
Harmon SA, Sanford TH, Xu S et al (2020) Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nat Commun 11(1):1–7
Prokop M, van Everdingen W, van Rees Vellinga T et al (2020) CO-RADS–A categorical CT assessment scheme for patients with suspected COVID-19: definition and evaluation. Radiology. 201473
Bernheim A (2020) Chest CT findings in COVID-19. Radiology. 19
Bellini D, Panvini N, Rengo M et al (2020) Diagnostic accuracy and interobserver variability of CO-RADS in patients with suspected coronavirus disease-2019: a multireader validation study. Eur Radiol:1–9
Lieveld AWE, Azijli K, Teunissen BP et al (2020) Chest CT in COVID-19 at the ED: validation of the COVID-19 reporting and data system (CORADS) and CT severity score. Chest 159(3):1126–1135
Grillet F, Behr J, Calame P, Aubry S, Delabrousse E (2020) Acute pulmonary embolism associated with COVID-19 pneumonia detected by pulmonary CT angiography. Radiology. 201544
Lang M, Som A, Mendoza DP et al (2020) Hypoxaemia related to COVID-19: vascular and perfusion abnormalities on dual-energy CT. Lancet Infect Dis 20(12):1365–1366
Funding
The authors state that this work has not received any funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Eduardo J. Mortani Barbosa Jr, MD.
Conflict of interest
The authors of this manuscript declare relationships with the following companies: Siemens AG.
Statistics and biometry
The following authors (EB, BG) have significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• multicenter study
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementarys information
ESM 1
(DOCX 1377 kb)
Rights and permissions
About this article
Cite this article
Mortani Barbosa, E.J., Georgescu, B., Chaganti, S. et al. Machine learning automatically detects COVID-19 using chest CTs in a large multicenter cohort. Eur Radiol 31, 8775–8785 (2021). https://doi.org/10.1007/s00330-021-07937-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-021-07937-3