Abstract
Objectives
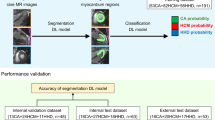
The high variability of hypertrophic cardiomyopathy (HCM) genetic phenotypes has prompted the establishment of risk-stratification systems that predict the risk of a positive genetic mutation based on clinical and echocardiographic profiles. This study aims to improve mutation-risk prediction by extracting cardiovascular magnetic resonance (CMR) morphological features using a deep learning algorithm.
Methods
We recruited 198 HCM patients (48% men, aged 47 ± 13 years) and divided them into training (147 cases) and test (51 cases) sets based on different genetic testing institutions and CMR scan dates (2012, 2013, respectively). All patients underwent CMR examinations, HCM genetic testing, and an assessment of established genotype scores (Mayo Clinic score I, Mayo Clinic score II, and Toronto score). A deep learning (DL) model was developed to classify the HCM genotypes, based on a nonenhanced four-chamber view of cine images.
Results
The areas under the curve (AUCs) for the test set were Mayo Clinic score I (AUC: 0.64, sensitivity: 64.29%, specificity: 47.83%), Mayo Clinic score II (AUC: 0.70, sensitivity: 64.29%, specificity: 65.22%), Toronto score (AUC: 0.74, sensitivity: 75.00%, specificity: 56.52%), and DL model (AUC: 0.80, sensitivity: 85.71%, specificity: 69.57%). The combination of the DL and the Toronto score resulted in a significantly higher predictive performance (AUC = 0.84, sensitivity: 83.33%, specificity: 78.26%), compared with Mayo I (p = 006), Mayo II (p = 022), and Toronto score (p = 0.029).
Conclusions
The combination of the DL model, based on nonenhanced cine CMR images and the Toronto score yielded significantly higher diagnostic performance in detecting HCM mutations.
Key Points
• Deep learning method could enable the extraction of image features from cine images.
• Deep learning method based on cine images performed better than established scores in identifying HCM patients with positive genotypes.
• The combination of the deep learning method based on cine images and the Toronto score could further improve the performance of the identification of HCM patients with positive genotypes.




Similar content being viewed by others
Abbreviations
- AUC:
-
Area under the (receiver-operating characteristic) curve
- CMR:
-
Cardiac magnetic resonance
- DL:
-
Deep learning
- FHHCM:
-
Family history of hypertrophic cardiomyopathy
- FHSCD:
-
Family history of sudden cardiac death
- HCM:
-
Hypertrophic cardiomyopathy
- LSTM:
-
Long short-term memory
- LVMWT:
-
Left ventricular maximal wall thickness
- LVPWT:
-
Left ventricular posterior wall thickness
- ROC:
-
Receiver-operating characteristic
- ROIs:
-
Regions of interest
- SCD:
-
sudden cardiac death
- VUS:
-
Variants of uncertain significance
References
Maron BJ (2002) Hypertrophic cardiomyopathy: a systematic review. JAMA 287:1308–1320
Maron BJ, Maron MS (2013) Hypertrophic cardiomyopathy. Lancet 381:242–255
Maron BJ, Maron MS, Semsarian C (2012) Genetics of hypertrophic cardiomyopathy after 20 years: clinical perspectives. J Am Coll Cardiol 60:705–715
Elliott PM, Anastasakis A, Borger MA et al (2014) 2014 ESC guidelines on diagnosis and management of hypertrophic cardiomyopathy: the task force for the diagnosis and management of hypertrophic cardiomyopathy of the European Society of Cardiology (ESC). Eur Heart J 35:2733–2779
Wordsworth S, Leal J, Blair E et al (2010) DNA testing for hypertrophic cardiomyopathy: a cost-effectiveness model. Eur Heart J 31:926–935
Van Driest SL, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ (2005) Sarcomeric genotyping in hypertrophic cardiomyopathy. Mayo Clin Proc 80:463–469
Andersen PS, Havndrup O, Hougs L et al (2009) Diagnostic yield, interpretation, and clinical utility of mutation screening of sarcomere encoding genes in Danish hypertrophic cardiomyopathy patients and relatives. Hum Mutat 30:363–370
Girolami F, Olivotto I, Passerini I et al (2006) A molecular screening strategy based on beta-myosin heavy chain, cardiac myosin binding protein C and troponin T genes in Italian patients with hypertrophic cardiomyopathy. J Cardiovasc Med (Hagerstown) 7:601–607
Lopes LR, Rahman MS, Elliott PM (2013) A systematic review and meta-analysis of genotype-phenotype associations in patients with hypertrophic cardiomyopathy caused by sarcomeric protein mutations. Heart 99:1800–1811
Lopes LR, Syrris P, Guttmann OP et al (2015) Novel genotype-phenotype associations demonstrated by high-throughput sequencing in patients with hypertrophic cardiomyopathy. Heart 101:294–301
Gruner C, Ivanov J, Care M et al (2013) Toronto hypertrophic cardiomyopathy genotype score for prediction of a positive genotype in hypertrophic cardiomyopathy. Circ Cardiovasc Genet 6:19–26
Li Y, Qian Z, Xu K et al (2018) MRI features predict p53 status in lower-grade gliomas via a machine-learning approach. Neuroimage Clin 17:306–311
Li Y, Liu X, Xu K et al (2018) MRI features can predict EGFR expression in lower grade gliomas: a voxel-based radiomic analysis. Eur Radiol 28:356–362
Zhang N, Yang G (2019) Deep learning for diagnosis of chronic myocardial infarction on nonenhanced cardiac cine MRI. Radiology 291:606–617
Baessler B, Luecke C, Lurz J et al (2018) Cardiac MRI texture analysis of T1 and T2 maps in patients with infarctlike acute myocarditis. Radiology 289:357–365
Cheng S, Fang M, Cui C et al (2018) LGE-CMR-derived texture features reflect poor prognosis in hypertrophic cardiomyopathy patients with systolic dysfunction: preliminary results. Eur Radiol 28:4615–4624
Bello GA, Dawes TJW, Duan J et al (2019) Deep learning cardiac motion analysis for human survival prediction. Nat Mach Intell 1:95–104
Bai W, Sinclair M, Tarroni G et al (2018) Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. J Cardiovasc Magn Reson 20:65
Bos JM, Will ML, Gersh BJ, Kruisselbrink TM, Ommen SR, Ackerman MJ (2014) Characterization of a phenotype-based genetic test prediction score for unrelated patients with hypertrophic cardiomyopathy. Mayo Clin Proc 89:727–737
Van Driest SL, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ (2005) Yield of genetic testing in hypertrophic cardiomyopathy. Mayo Clin Proc 80:739–744
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–424
Binder J, Ommen SR, Gersh BJ et al (2006) Echocardiography-guided genetic testing in hypertrophic cardiomyopathy: septal morphological features predict the presence of myofilament mutations. Mayo Clin Proc 81:459–467
Chen LC, Zhu Y, Papandreou G, Schroff F, Adam H (2018) Encoder-decoder with atrous separable convolution for semantic image segmentation. European Conference on Computer Vision (ECCV), Munich. Available via https://doi.org/10.1007/978-3-030-01234-2_49. Accessed 06 October 2018
Szegedy C, Ioffe S, Vanhoucke VAlemi A (2016) Inception-v4, Inception-ResNet and the impact of residual connections on learning. National Conference on Artificial Intelligence(AAAI) , Phoenix. Available via arXiv:1602.07261. Accessed: February 2016
Deng J, Dong W, Socher R, Li LJ, Li K, Li FF (2009) ImageNet: a large-scale hierarchical image database. Computer Vision & Pattern Recognition (CVPR), Miami. Available via https://doi.org/10.1109/CVPR.2009.5206848. Accessed 18 August 2009
Gers FA, Schmidhuber J, Cummins F (2000) Learning to forget: continual prediction with LSTM. Neural Comput 12:2451–2471
Kingma DP, Ba J (2015) Adam: a method for stochastic optimization. International Conference on Learning Representations (ICLR), San Diego. Available via arXiv:1412.6980. Accessed 2015
Chollet F (2018) Keras: the python deep learning library. Astrophysics Source Code Library. Available via https://keras.io/getting-started/faq/#how-should-i-cite-keras. Accessed June 2018
Bos JM, Towbin JA, Ackerman MJ (2009) Diagnostic, prognostic, and therapeutic implications of genetic testing for hypertrophic cardiomyopathy. J Am Coll Cardiol 54:201–211
Baessler B, Mannil M, Oebel S, Maintz D, Alkadhi H, Manka R (2018) Subacute and chronic left ventricular myocardial scar: accuracy of texture analysis on nonenhanced cine MR images. Radiology 286:103–112
Tao Q, Yan W, Wang Y et al (2019) Deep learning-based method for fully automatic quantification of left ventricle function from cine MR images: a multivendor, multicenter study. Radiology 290:81–88
Ellims AH, Iles LM, Ling LH et al (2014) A comprehensive evaluation of myocardial fibrosis in hypertrophic cardiomyopathy with cardiac magnetic resonance imaging: linking genotype with fibrotic phenotype. Eur Heart J Cardiovasc Imaging 15:1108–1116
Fujita T, Fujino N, Anan R et al (2013) Sarcomere gene mutations are associated with increased cardiovascular events in left ventricular hypertrophy: results from multicenter registration in Japan. JACC Heart Fail 1:459–466
Olivotto I, Girolami F, Ackerman MJ et al (2008) Myofilament protein gene mutation screening and outcome of patients with hypertrophic cardiomyopathy. Mayo Clin Proc 83:630–638
Funding
This paper is supported by the National Natural Science Foundation of China under Grant Nos. 81922040, 81930053, 81227901, 81527805, and 81772012; the major international (regional) joint research project of National Science Foundation of China under Grant No. 81620108015; Beijing Natural Science Foundation (under Grant No. 7182109); National Key Research and Development Plan of China (under Grant Nos. 2017YFA0205200, 2016YFA0100900, and 2016YFA0100902); and Youth Innovation Promotion Association CAS (Grant No. 2019136).
Author information
Authors and Affiliations
Contributions
Lu Li made contributions to the conception and design of the study; Lu Li and Hongyu Zhou drafted the manuscript; Kankan Zhao and Lu Li were responsible for statistical analysis of the data, Zhenyu Liu, Min-jie Lu, and Xiuyu Chen made critical revisions to the manuscript; Lu Li and Lei Song collected conventional CMR and genetic data; Gang Yin was in charge of image segmentation; Shihua zhao, Jie Tian, and Hairong Zheng made a contribution to study conduction. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Shihua Zhao.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was obtained from all subjects (patients) in this study.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• Retrospective
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 21.3 kb)
Rights and permissions
About this article
Cite this article
Zhou, H., Li, L., Liu, Z. et al. Deep learning algorithm to improve hypertrophic cardiomyopathy mutation prediction using cardiac cine images. Eur Radiol 31, 3931–3940 (2021). https://doi.org/10.1007/s00330-020-07454-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07454-9