Abstract
Objective
To evaluate machine learning–based classifiers in detecting clinically significant prostate cancer (PCa) with Prostate Imaging Reporting and Data System (PI-RADS) score 3 lesions.
Methods
We retrospectively enrolled 346 patients with PI-RADS 3 lesions at two institutions. All patients underwent prostate multiparameter MRI (mpMRI) and transperineal MRI-ultrasonography (MRI-US)-targeted biopsy. We collected data on age, pre-biopsy serum prostate-specific antigen (PSA) level, prostate volume (PV), PSA density (PSAD), the location of suspicious PI-RADS 3 lesions, and histopathology results. Four machine learning–based classifiers—logistic regression, support vector machine, eXtreme Gradient Boosting (XGBoost), and random forest—were trained using datasets from Nanjing Drum Tower Hospital. External validation was carried out using datasets from Molinette Hospital.
Results
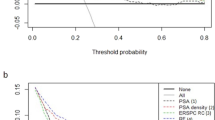
Among 287 PI-RADS 3 patients, prostate cancer was proven pathologically in 59 (20.6%), and 228 (79.4%) had benign lesions. For 380 PI-RADS 3 lesions, 81 (21.3%) were proven to be PCa and 299 (78.7%) benign. Among four classifiers, the random forest classifier had the best performance in both patient-based and lesion-based datasets, with overall accuracy of 0.713 and 0.860, sensitivity of 0.857 and 0.613, and area under curve (AUC) of 0.771 and 0.832, respectively. In external validation, our best classifiers had an AUC of 0.688 with the best sensitivity (0.870) and specificity (0.500) in the 59 PI-RADS 3 patients in Molinette Hospital dataset.
Conclusions
The machine learning–based random forest classifier provided a reliable probability if a PI-RADS 3 patient was benign.
Key Points
• Machine learning–based classifiers could combine the clinical characteristics with accessible information on image report of PI-RADS 3 patient to generate a probability of malignancy.
• This probability could assist surgeons to make diagnostic decisions with more confidence and higher efficiency.






Similar content being viewed by others
Abbreviations
- ADC:
-
Apparent diffusion coefficient
- AI:
-
Artificial intelligence
- AP:
-
Anterior-posterior
- AUC:
-
Area under curve
- CDR:
-
Cancer detection rate
- DWI:
-
Diffusion-weighted imaging
- FH:
-
Foot-head
- mpMRI:
-
Multiparameter MRI
- MRI-US:
-
MRI-ultrasonography
- PCa:
-
Prostate cancer
- PI-RADS:
-
Prostate Imaging Reporting and Data System
- PSA:
-
Prostate-specific antigen
- PSAD:
-
Prostate-specific antigen density
- PV:
-
Prostate volume
- RL:
-
Right-left
- ROC:
-
Receiver operating characteristic
- SVM:
-
Support vector machine
- T2W:
-
T2-weighted
- XGBoost:
-
eXtreme Gradient Boosting
References
Wong MC, Goggins WB, Wang HH et al (2016) Global incidence and mortality for prostate cancer: analysis of temporal patterns and trends in 36 countries. Eur Urol 70(5):862–874
Chen W, Zheng R, Baade PD et al (2016) Cancer statistics in China, 2015. CA Cancer J Clin 66(2):115–132
Barentsz JO, Weinreb JC, Verma S et al (2016) Synopsis of the PI-RADS v2 guidelines for multiparametric prostate magnetic resonance imaging and recommendations for use. Eur Urol 69(1):41–49
Ahmed HU, El-Shater Bosaily A, Brown LC et al (2017) Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet 389(10071):815–822
van der Leest M, Cornel E, Israel B et al (2019) Head-to-head comparison of transrectal ultrasound-guided prostate biopsy versus multiparametric prostate resonance imaging with subsequent magnetic resonance-guided biopsy in biopsy-naive men with elevated prostate-specific antigen: a large prospective multicenter clinical study. Eur Urol 75(4):570–578
Venderink W, van Luijtelaar A, Bomers JG et al (2017) Results of targeted biopsy in men with magnetic resonance imaging lesions classified equivocal, likely or highly likely to be clinically significant prostate cancer. Eur Urol 73(3):353–360
Hermie I, Van Besien J, De Visschere P, Lumen N, Decaestecker K (2019) Which clinical and radiological characteristics can predict clinically significant prostate cancer in PI-RADS 3 lesions? A retrospective study in a high-volume academic center. Eur J Radiol 114:92–98
Bektas CT, Kocak B, Yardimci AH et al (2019) Clear cell renal cell carcinoma: machine learning-based quantitative computed tomography texture analysis for prediction of Fuhrman nuclear grade. Eur Radiol 29(3):1153–1163
Yao L, Cai M, Chen Y, Shen C, Shi L, Guo Y (2019) Prediction of antiepileptic drug treatment outcomes of patients with newly diagnosed epilepsy by machine learning. Epilepsy Behav 96:92–97
Ambale-Venkatesh B, Yang X, Wu CO et al (2017) Cardiovascular event prediction by machine learning: the multi-ethnic study of atherosclerosis. Circ Res 121(9):1092–1101
Schelb P, Kohl S, Radtke JP et al (2019) Classification of cancer at prostate MRI: deep learning versus clinical PI-RADS assessment. Radiology 293(3):607–617
Huang H, Wang W, Lin T et al (2016) Comparison of the complications of traditional 12 cores transrectal prostate biopsy with image fusion guided transperineal prostate biopsy. BMC Urol 16(1):68
Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA (2016) The 2014 International Society of Urological Pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma: definition of grading patterns and proposal for a new grading system. Am J Surg Pathol 40(2):244–252
Fabian Pedregosa GV, Gramfort A, Michel V et al (2011) Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830
Tolles J, Meurer WJ (2016) Logistic regression: relating patient characteristics to outcomes. JAMA 316(5):533–534
Huang S, Cai N, Pacheco PP, Narrandes S, Wang Y, Xu W (2018) Applications of support vector machine (SVM) learning in cancer genomics. Cancer Genomics Proteomics 15(1):41–51
Ogunleye AA, Qing-Guo W (2019) XGBoost model for chronic kidney disease diagnosis. IEEE/ACM Trans Comput Biol Bioinform. https://doi.org/10.1109/TCBB.2019.2911071
Breiman L (2001) Random forests. Mach Learn 45(1):5–32
Pavey TG, Gilson ND, Gomersall SR, Clark B, Trost SG (2017) Field evaluation of a random forest activity classifier for wrist-worn accelerometer data. J Sci Med Sport 20(1):75–80
Zweig MH, Campbell G (1993) Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem 39(4):561–577
Ruuska S, Hamalainen W, Kajava S, Mughal M, Matilainen P, Mononen J (2018) Evaluation of the confusion matrix method in the validation of an automated system for measuring feeding behaviour of cattle. Behav Processes 148:56–62
Hansen NL, Koo BC, Warren AY, Kastner C, Barrett T (2017) Sub-differentiating equivocal PI-RADS-3 lesions in multiparametric magnetic resonance imaging of the prostate to improve cancer detection. Eur J Radiol 95:307–313
Sheridan AD, Nath SK, Syed JS et al (2018) Risk of clinically significant prostate cancer associated with prostate imaging reporting and data system category 3 (equivocal) lesions identified on multiparametric prostate MRI. AJR Am J Roentgenol 210(2):347–357
Funding
This work was supported by grants from the National Natural Science Foundation of China (81602221) and the National Natural Science Foundation of Jiangsu Province (BK20160117).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Hongqian Guo.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors has significant statistical expertise.
Ethical approval
Institutional review board approval was not required because patient data was not so detailed as to raise privacy concerns and the analysis was retrospective.
Informed consent
Written informed consent was waived by the institutional review board.
Methodology
• Retrospective
• Diagnostic study
• Multicenter study
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 2976 kb)
Appendix
Appendix
XGBoost is based on gradient tree boosting, an algorithm with which new models are created that predict the residuals of prior models and are then added together for the final prediction [17].
In random forest, each tree was developed from a bootstrap sample of the training dataset, and each node was the part that was the best among a haphazard-chosen subset of features. The class predictions created by each tree within the forest were amassed, and the ultimate prediction was based on the lion’s share vote [18, 19].
Rights and permissions
About this article
Cite this article
Kan, Y., Zhang, Q., Hao, J. et al. Clinico-radiological characteristic-based machine learning in reducing unnecessary prostate biopsies of PI-RADS 3 lesions with dual validation. Eur Radiol 30, 6274–6284 (2020). https://doi.org/10.1007/s00330-020-06958-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-06958-8