Abstract
Objectives
To establish a quantitative MR model that uses clinically relevant features of tumor location and tumor volume to differentiate lower grade glioma (LRGG, grades II and III) and glioblastoma (GBM, grade IV).
Methods
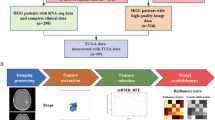
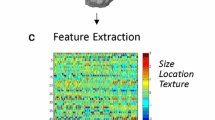
We extracted tumor location and tumor volume (enhancing tumor, non-enhancing tumor, peritumor edema) features from 229 The Cancer Genome Atlas (TCGA)-LGG and TCGA-GBM cases. Through two sampling strategies, i.e., institution-based sampling and repeat random sampling (10 times, 70% training set vs 30% validation set), LASSO (least absolute shrinkage and selection operator) regression and nine–machine learning method–based models were established and evaluated.
Results
Principal component analysis of 229 TCGA-LGG and TCGA-GBM cases suggested that the LRGG and GBM cases could be differentiated by extracted features. For nine machine learning methods, stack modeling and support vector machine achieved the highest performance (institution-based sampling validation set, AUC > 0.900, classifier accuracy > 0.790; repeat random sampling, average validation set AUC > 0.930, classifier accuracy > 0.850). For the LASSO method, regression model based on tumor frontal lobe percentage and enhancing and non-enhancing tumor volume achieved the highest performance (institution-based sampling validation set, AUC 0.909, classifier accuracy 0.830). The formula for the best performance of the LASSO model was established.
Conclusions
Computer-generated, clinically meaningful MRI features of tumor location and component volumes resulted in models with high performance (validation set AUC > 0.900, classifier accuracy > 0.790) to differentiate lower grade glioma and glioblastoma.
Key Points
• Lower grade glioma and glioblastoma have significant different location and component volume distributions.
• We built machine learning prediction models that could help accurately differentiate lower grade gliomas and GBM cases. We introduced a fast evaluation model for possible clinical differentiation and further analysis.



Similar content being viewed by others
Change history
09 March 2020
The original version of this article, published on 05 February 2020, unfortunately contained a mistake.
Abbreviations
- AUC:
-
Area under ROC
- BraTS:
-
Multimodal Brain Tumor Segmentation Challenge
- CA:
-
Classification accuracy
- FLAIR:
-
Fluid-attenuated inversion recovery
- FMRIB:
-
Functional magnetic resonance imaging of the brain
- FSL:
-
FMRIB Software Library
- GBM:
-
Glioblastoma
- IG:
-
Information gain
- KNN:
-
k-nearest neighbors algorithm
- LASSO:
-
Least absolute shrinkage and selection operator
- LGG:
-
Low-grade glioma
- LRGG:
-
Lower grade glioma
- MNI:
-
Montreal Neurological Institute
- NIfTI:
-
Neuroimaging Informatics Technology Initiative
- PCs:
-
Principal components
- PCA:
-
Principal component analysis
- RF:
-
Random forest
- SVM:
-
Support vector machine
- TCGA:
-
The Cancer Genome Atlas
- TCIA:
-
The Cancer Imaging Archive
- Volume_ED:
-
Peritumoral edema tumor volume
- Volume_ET:
-
Enhancing tumor volume
- Volume_NET:
-
Non-enhancing tumor volume
- Volume_TC:
-
Tumor core volume
- Volume_WT:
-
Whole tumor volume
References
Schiff D, Van den Bent M, Vogelbaum MA et al (2019) Recent developments and future directions in adult lower-grade gliomas: Society for Neuro-Oncology (SNO) and European Association of Neuro-Oncology (EANO) consensus. Neuro Oncol 21:837–853
Asari S, Makabe T, Katayama S, Itoh T, Tsuchida S, Ohmoto T (1994) Assessment of the pathological grade of astrocytic gliomas using an MRI score. Neuroradiology 36:308–310
Pierallini A, Bonamini M, Bozzao A et al (1997) Supratentorial diffuse astrocytic tumours: proposal of an MRI classification. Eur Radiol 7:395–399
Cho H-H, Park H (2017) Classification of low-grade and highgrade glioma using multi-modal image radiomics features. Conf Proc IEEE Eng Med Biol Soc 2017:3081–3084
Togao O, Hiwatashi A, Yamashita K et al (2015) Differentiation of high-grade and low-grade diffuse gliomas by intravoxel incoherent motion MR imaging. Neuro Oncol 18:132–141
Rizzo S, Botta F, Raimondi S et al (2018) Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp 2:36
Larjavaara S, Mäntylä R, Salminen T et al (2007) Incidence of gliomas by anatomic location. Neuro Oncol 9:319–325
Carlson MR, Pope WB, Horvath S et al (2007) Relationship between survival and edema in malignant gliomas: role of vascular endothelial growth factor and neuronal pentraxin 2. Clin Cancer Res 13:2592–2598
Carrillo J, Lai A, Nghiemphu P et al (2012) Relationship between tumor enhancement, edema, IDH1 mutational status, MGMT promoter methylation, and survival in glioblastoma. AJNR Am J Neuroradiol 33:1349–1355
Clark K, Vendt B, Smith K et al (2013) The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 26:1045–1057
Koçak B, Durmaz ES, Ates E, Kilickesmez O (2019) Radiomics with artificial intelligence: a practical guide for beginners. Diagn Interv Radiol. https://doi.org/10.5152/dir.2019.19321
Bakas S, Zeng K, Sotiras A et al (2015) GLISTRboost: combining multimodal MRI segmentation, registration, and biophysical tumor growth modeling with gradient boosting machines for glioma segmentation. Brainlesion 9556:144–155
Grabner G, Janke AL, Budge MM, Smith D, Pruessner J, Collins DL (2006) Symmetric atlasing and model based segmentation: an application to the hippocampus in older adults. In: Larsen R, Nielsen M, Sporring J (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2006.Lecture Notes in Computer Science, vol 4191. Springer, Berlin, Heidelberg
Woolrich MW, Jbabdi S, Patenaude B et al (2009) Bayesian analysis of neuroimaging data in FSL. Neuroimage 45:S173–S186
Mazziotta J, Toga A, Evans A et al (2001) A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos Trans R Soc Lond B Biol Sci 356:1293–1322
Bakas S, Akbari H, Sotiras A et al (2017) Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data 4:170117
Demšar J, Curk T, Erjavec A et al (2013) Orange: data mining toolbox in Python. J Mach Learn Res 14:2349–2353
Piotr Romanski LK (2018) Package FSelector - CRAN. CRAN. Available via https://cran.r-project.org/web/packages/FSelector/index.html. Accessed Oct 22 2019
Friedman J, Hastie T, Tibshirani R (2010) Regularization paths for generalized linear models via coordinate descent. J Stat Softw 33:1
Kuhn M (2008) Building predictive models in R using the caret package. J Stat Softw 28:1–26
Robin X, Turck N, Hainard A et al (2011) pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12:77
Hsieh KL-C, Lo C-M, Hsiao C-J (2017) Computer-aided grading of gliomas based on local and global MRI features. Comput Methods Programs Biomed 139:31–38
Stroebe H (1895) Uber entstehung und bau der gehirngliome. Beitr Pathol Anat Allg Pathol 18:405–485
Burkhard C, Di Patre P-L, Schüler D et al (2003) A population-based study of the incidence and survival rates in patients with pilocytic astrocytoma. J Neurosurg 98:1170–1174
Hargrave D, Bartels U, Bouffet E (2006) Diffuse brainstem glioma in children: critical review of clinical trials. Lancet Oncol 7:241–248
Pallud J, Capelle L, Taillandier L et al (2009) Prognostic significance of imaging contrast enhancement for WHO grade II gliomas. Neuro Oncol 11:176–182
Suchorska B, Schüller U, Biczok A et al (2019) Contrast enhancement is a prognostic factor in IDH1/2 mutant, but not in wild-type WHO grade II/III glioma as confirmed by machine learning. Eur J Cancer 107:15–27
Wang Y, Wang K, Li S et al (2015) Patterns of tumor contrast enhancement predict the prognosis of anaplastic gliomas with IDH1 mutation. AJNR Am J Neuroradiol 36:2023–2029
Scott J, Brasher P, Sevick R, Rewcastle N, Forsyth P (2002) How often are nonenhancing supratentorial gliomas malignant? A population study. Neurology 59:947–949
Reyes-Botero G, Dehais C, Idbaih A et al (2013) Contrast enhancement in 1p/19q-codeleted anaplastic oligodendrogliomas is associated with 9p loss, genomic instability, and angiogenic gene expression. Neuro Oncol 16:662–670
Acknowledgments
The results here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Funding
The authors state that this work has not received any funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Dr. Robert Fulbright.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was not required because only deidentified data is used in this study.
Study subjects or cohorts overlap
Some study subjects or cohorts have been previously reported. The TCGA-LGG and TCGA-GBM cohorts were involved in this study. Due to the enormous publishing scale, here we are not able to list or include every associated study. However, the cohort resource and include/exclude criteria have been introduced and cited in detail.
The major differences between the cohort used in our study and other study are as follows:
1. In our study, only TCGA-LGG and TCGA-GBM cases with MRI images available on TCIA were included.
2. The abovementioned cases were subsequently selected by segmentation data availability; only cases that have GLISTRboost-generated and manually checked segmentation data were selected.
Methodology
• retrospective
• diagnostic study
• multicenter study
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised: The name of E. Zeynep Erson-Omay was presented incorrectly.
Rights and permissions
About this article
Cite this article
Cao, H., Erson-Omay, E.Z., Li, X. et al. A quantitative model based on clinically relevant MRI features differentiates lower grade gliomas and glioblastoma. Eur Radiol 30, 3073–3082 (2020). https://doi.org/10.1007/s00330-019-06632-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-019-06632-8