Abstract
Objectives
To develop a proof-of-concept “interpretable” deep learning prototype that justifies aspects of its predictions from a pre-trained hepatic lesion classifier.
Methods
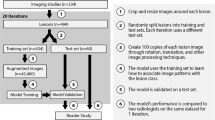
A convolutional neural network (CNN) was engineered and trained to classify six hepatic tumor entities using 494 lesions on multi-phasic MRI, described in Part 1. A subset of each lesion class was labeled with up to four key imaging features per lesion. A post hoc algorithm inferred the presence of these features in a test set of 60 lesions by analyzing activation patterns of the pre-trained CNN model. Feature maps were generated that highlight regions in the original image that correspond to particular features. Additionally, relevance scores were assigned to each identified feature, denoting the relative contribution of a feature to the predicted lesion classification.
Results
The interpretable deep learning system achieved 76.5% positive predictive value and 82.9% sensitivity in identifying the correct radiological features present in each test lesion. The model misclassified 12% of lesions. Incorrect features were found more often in misclassified lesions than correctly identified lesions (60.4% vs. 85.6%). Feature maps were consistent with original image voxels contributing to each imaging feature. Feature relevance scores tended to reflect the most prominent imaging criteria for each class.
Conclusions
This interpretable deep learning system demonstrates proof of principle for illuminating portions of a pre-trained deep neural network’s decision-making, by analyzing inner layers and automatically describing features contributing to predictions.
Key Points
• An interpretable deep learning system prototype can explain aspects of its decision-making by identifying relevant imaging features and showing where these features are found on an image, facilitating clinical translation.
• By providing feedback on the importance of various radiological features in performing differential diagnosis, interpretable deep learning systems have the potential to interface with standardized reporting systems such as LI-RADS, validating ancillary features and improving clinical practicality.
• An interpretable deep learning system could potentially add quantitative data to radiologic reports and serve radiologists with evidence-based decision support.




Similar content being viewed by others
Abbreviations
- CNN:
-
Convolutional neural network
- CRC:
-
Colorectal carcinoma
- DL:
-
Deep learning
- FNH:
-
Focal nodular hyperplasia
- HCC:
-
Hepatocellular carcinoma
- ICC:
-
Intrahepatic cholangiocarcinoma
- LI-RADS:
-
Liver Imaging Reporting and Data System
- PPV:
-
Positive predictive value
- Sn:
-
Sensitivity
References
Rajpurkar P, Irvin J, Zhu K et al (2018) Deep learning for chest radiograph diagnosis: a retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med 15:e1002686. https://doi.org/10.1371/journal.pmed.1002686
Chartrand G, Cheng PM, Vorontsov E et al (2017) Deep learning: a primer for radiologists. Radiographics 37:2113–2131
Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou S (2016) Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imaging 35:1207–1216
Greenspan H, Van Ginneken B, Summers RM (2016) Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans Med Imaging 35:1153–1159
Hamm CA, Wang CJ, Savic LJ et al (2019) Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol. https://doi.org/10.1007/s00330-019-06205-9
Olden JD, Jackson DA (2002) Illuminating the “black box”: a randomization approach for understanding variable contributions in artificial neural networks. Ecol Model 154:135–150
Kiczales G (1996) Beyond the black box: open implementation. IEEE Softw 13(8):10–11
Dayhoff JE, DeLeo JM (2001) Artificial neural networks: opening the black box. Cancer 91:1615–1635
Olah C, Satyanarayan A, Johnson I et al (2018) The building blocks of interpretability. Distill 3:e10. https://doi.org/10.23915/distill.00010
Corwin MT, Lee AY, Fananapazir G, Loehfelm TW, Sarkar S, Sirlin CB (2018) Nonstandardized terminology to describe focal liver lesions in patients at risk for hepatocellular carcinoma: implications regarding clinical communication. AJR Am J Roentgenol 210:85–90
Mitchell DG, Bruix J, Sherman M, Sirlin CB (2015) LI-RADS (Liver Imaging Reporting and Data System): summary, discussion, and consensus of the LI-RADS Management Working Group and future directions. Hepatology 61:1056–1065
Mitchell DG, Bashir MR, Sirlin CB (2018) Management implications and outcomes of LI-RADS-2, -3, -4, and -M category observations. Abdom Radiol (NY) 43:143–148
Barth B, Donati O, Fischer M et al (2016) Reliability, validity, and reader acceptance of LI-RADS-an in-depth analysis. Acad Radiol 23:1145
Davenport MS, Khalatbari S, Liu PS et al (2014) Repeatability of diagnostic features and scoring systems for hepatocellular carcinoma by using MR imaging. Radiology 272:132
Ehman EC, Behr SC, Umetsu SE et al (2016) Rate of observation and inter-observer agreement for LI-RADS major features at CT and MRI in 184 pathology proven hepatocellular carcinomas. Abdom Radiol (NY) 41:963–969
Zhang YD, Zhu FP, Xu X et al (2016) Classifying CT/MR findings in patients with suspicion of hepatocellular carcinoma: comparison of liver imaging reporting and data system and criteria-free Likert scale reporting models. J Magn Reson Imaging 43:373–383
Bashir M, Huang R, Mayes N et al (2015) Concordance of hypervascular liver nodule characterization between the organ procurement and transplant network and liver imaging reporting and data system classifications. J Magn Reson Imaging 42:305
Liu W, Qin J, Guo R et al (2017) Accuracy of the diagnostic evaluation of hepatocellular carcinoma with LI-RADS. Acta Radiol. https://doi.org/10.1177/0284185117716700:284185117716700
Fowler KJ, Tang A, Santillan C et al (2018) Interreader reliability of LI-RADS version 2014 algorithm and imaging features for diagnosis of hepatocellular carcinoma: a large international multireader study. Radiology 286:173–185
Cruite I, Santillan C, Mamidipalli A, Shah A, Tang A, Sirlin CB (2016) Liver imaging reporting and data system: review of ancillary imaging features. Semin Roentgenol 51:301–307. https://doi.org/10.1053/j.ro.2016.05.004
Sirlin CB, Kielar AZ, Tang A, Bashir MR (2018) LI-RADS: a glimpse into the future. Abdom Radiol (NY) 43:231–236
Kim YY, An C, Kim S, Kim MJ (2017) Diagnostic accuracy of prospective application of the Liver Imaging Reporting and Data System (LI-RADS) in gadoxetate-enhanced MRI. Eur Radiol. https://doi.org/10.1007/s00330-017-5188-y
Molnar C (2019) Interpretable machine learning. A guide for making black box models explainable. https://christophm.github.io/interpretable-ml-book/
Fisher A, Rudin C, Dominici F (2018) Model class reliance: variable importance measures for any machine learning model class, from the “Rashomon” perspective. arXiv preprint arXiv:180101489
Federle MP, Jeffrey RB, Woodward PJ, Borhani A (2009) Diagnostic imaging: abdomen. Published by Amirsys. Lippincott Williams & Wilkins
Victoria C, Sirlin CB, Cui J et al (2018) LI-RADS v2018 CT/MRI Manual. Available via https://www.acr.org/-/media/ACR/Files/Clinical-Resources/LIRADS/Chapter-16-Imaging-features.pdf?la=en
Everitt BS (2002) The Cambridge dictionary of statistics. Cambridge University Press
Koh PW, Liang P (2017) Understanding black-box predictions via influence functions. AarXiv preprint arXiv:170304730
Narsinh KH, Cui J, Papadatos D, Sirlin CB, Santillan CS (2018) Hepatocarcinogenesis and LI-RADS. Abdom Radiol (NY) 43:158–168
Tang A, Bashir MR, Corwin MT et al (2018) Evidence supporting LI-RADS major features for CT- and MR imaging-based diagnosis of hepatocellular carcinoma: a systematic review. Radiology 286:29–48
Holzinger A, Biemann C, Pattichis CS, Kell DB (2017) What do we need to build explainable AI systems for the medical domain? arXiv preprint arXiv:171209923
Funding
BL and CW received funding from the Radiological Society of North America (RSNA Research Resident Grant No. RR1731). JD, JC, ML, and CW received funding from the National Institutes of Health (NIH/NCI R01 CA206180).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Julius Chapiro.
Conflict of interest
The authors of this manuscript declare relationships with the following companies: JW: Bracco Diagnostics, Siemens AG; ML: Pro Medicus Limited; JC: Koninklijke Philips, Guerbet SA, Eisai Co.
Statistics and biometry
One of the authors has significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• experimental
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 28.6 kb)
Rights and permissions
About this article
Cite this article
Wang, C.J., Hamm, C.A., Savic, L.J. et al. Deep learning for liver tumor diagnosis part II: convolutional neural network interpretation using radiologic imaging features. Eur Radiol 29, 3348–3357 (2019). https://doi.org/10.1007/s00330-019-06214-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-019-06214-8