Abstract
Purpose
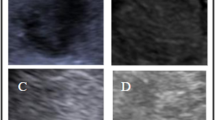
The ability to reliably distinguish benign from malignant solid liver lesions on ultrasonography can increase access, decrease costs, and help to better triage patients for biopsy. In this study, we used deep learning to differentiate benign from malignant focal solid liver lesions based on their ultrasound appearance.
Methods
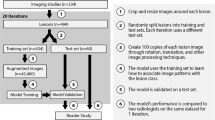
Among the 596 patients who met the inclusion criteria, there were 911 images of individual liver lesions, of which 535 were malignant and 376 were benign. Our training set contained 660 lesions augmented dynamically during training for a total of 330,000 images; our test set contained 79 images. A neural network with ResNet50 architecture was fine-tuned using pre-trained weights on ImageNet. Non-cystic liver lesions with definite diagnosis by histopathology or MRI were included. Accuracy of the final model was compared with expert interpretation. Two separate datasets were used in training and evaluation, one with all lesions and one with lesions deemed to be of uncertain diagnosis based on the Code Abdomen rating system.
Results
Our model trained on the complete set of all lesions achieved a test accuracy of 0.84 (95% CI 0.74–0.90) compared to expert 1 with a test accuracy of 0.80 (95% CI 0.70–0.87) and expert 2 with a test accuracy of 0.73 (95% CI 0.63–0.82). Our model trained on the uncertain set of lesions achieved a test accuracy of 0.79 (95% CI 0.69–0.87) compared to expert 1 with a test accuracy of 0.70 (95% CI 0.59–0.78) and expert 2 with a test accuracy of 0.66 (95% CI 0.55–0.75). On the uncertain dataset, compared to all experts averaged, the model had higher test accuracy (0.79 vs. 0.68, p = 0.025).
Conclusion
Deep learning algorithms proposed in the current study improve differentiation of benign from malignant ultrasound-captured solid liver lesions and perform comparably to expert radiologists. Deep learning tools can potentially be used to improve the accuracy and efficiency of clinical workflows.




Similar content being viewed by others
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- HCC:
-
Hepatocellular carcinoma
- ResNet:
-
Residual Network
- AUC:
-
Area under the curve
- ROC:
-
Receiver operating characteristic curve
- t-SNE:
-
T-Distributed stochastic neighbor embedding
- SVM:
-
Support vector machine
References
Bonder A, Afdhal N (2012) Evaluation of liver lesions. Clinics in liver disease 16:271–283
Toro A, Mahfouz A-E, Ardiri A, et al (2014) What is changing in indications and treatment of hepatic hemangiomas. A review. Annals of hepatology 13:327–339
Marrero JA, Ahn J, Reddy KR (2014) ACG clinical guideline: The diagnosis and management of focal liver lesions. The American journal of gastroenterology 109:1328
Dietrich CF, Kratzer W, Strobel D, et al (2006) Assessment of metastatic liver disease in patients with primary extrahepatic tumors by contrast-enhanced sonography versus ct and mri. World Journal of Gastroenterology: WJG 12:1699
Semelka RC, Martin DR, Balci C, Lance T (2001) Focal liver lesions: Comparison of dual-phase ct and multisequence multiplanar mr imaging including dynamic gadolinium enhancement. Journal of Magnetic Resonance Imaging: An Official Journal of the International Society for Magnetic Resonance in Medicine 13:397–401
Harvey CJ, Albrecht T (2001) Ultrasound of focal liver lesions. European radiology 11:1578–1593
Sodickson A, Baeyens PF, Andriole KP, et al (2009) Recurrent ct, cumulative radiation exposure, and associated radiation-induced cancer risks from ct of adults. Radiology 251:175–184
Ginde AA, Foianini A, Renner DM, et al (2008) Availability and quality of computed tomography and magnetic resonance imaging equipment in us emergency departments. Academic emergency medicine 15:780–783
Stevens MA, McCullough PA, Tobin KJ, et al (1999) A prospective randomized trial of prevention measures in patients at high risk for contrast nephropathy: Results of the prince study. Journal of the American College of Cardiology 33:403–411
Perez-Rodriguez J, Lai S, Ehst BD, et al (2009) Nephrogenic systemic fibrosis: Incidence, associations, and effect of risk factor assessment—report of 33 cases. Radiology 250:371–377
Thampanitchawong P, Piratvisuth T (1999) Liver biopsy: Complications and risk factors. World journal of gastroenterology 5:301
Sherman M, Peltekian KM, Lee C (1995) Screening for hepatocellular carcinoma in chronic carriers of hepatitis b virus: Incidence and prevalence of hepatocellular carcinoma in a north american urban population. Hepatology 22:432–438
Esteva A, Kuprel B, Novoa RA, et al (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542:115
Lakhani P, Sundaram B (2017) Deep learning at chest radiography: Automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology 284:574–582
Brown JM, Campbell JP, Beers A, et al (2018) Automated diagnosis of plus disease in retinopathy of prematurity using deep convolutional neural networks. JAMA ophthalmology
Gulshan V, Peng L, Coram M, et al (2016) Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. Jama 316:2402–2410
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the ieee conference on computer vision and pattern recognition. pp 770–778
Yasaka K, Akai H, Abe O, Kiryu S (2017) Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced ct: A preliminary study. Radiology 286:887–896
Zafar HM, Chadalavada SC, Kahn Jr CE, et al (2015) Code abdomen: An assessment coding scheme for abdominal imaging findings possibly representing cancer. Journal of the American College of Radiology: JACR 12:947
Strauss E, de Ferreira A SP, França AVC, et al (2015) Diagnosis and treatment of benign liver nodules: Brazilian society of hepatology (sbh) recommendations. Arquivos de gastroenterologia 52:47–54
Anderson SW, Kruskal JB, Kane RA (2009) Benign hepatic tumors and iatrogenic pseudotumors. Radiographics 29:211–229
Qian H, Li S, Ji M, Lin G (2016) MRI characteristics for the differential diagnosis of benign and malignant small solitary hypovascular hepatic nodules. European journal of gastroenterology & hepatology 28:749
Albiin N (2012) MRI of focal liver lesions. Current medical imaging reviews 8:107–116
Fowler KJ, Brown JJ, Narra VR (2011) Magnetic resonance imaging of focal liver lesions: Approach to imaging diagnosis. Hepatology 54:2227–2237
Itai Y, Ohtomo K, Furui S, et al (1985) Noninvasive diagnosis of small cavernous hemangioma of the liver: Advantage of mri. American journal of roentgenology 145:1195–1199
Willatt JM, Hussain HK, Adusumilli S, Marrero JA (2008) MR imaging of hepatocellular carcinoma in the cirrhotic liver: Challenges and controversies. Radiology 247:311–330
Chang K, Bai HX, Zhou H, et al (2018) Residual convolutional neural network for the determination of idh status in low-and high-grade gliomas from mr imaging. Clinical Cancer Research 24:1073–1081
Deng J, Dong W, Socher R, et al (2009) Imagenet: A large-scale hierarchical image database. In: Computer vision and pattern recognition, 2009. CVPR 2009. IEEE conference on. Ieee, pp 248–255
Maaten L van der, Hinton G (2008) Visualizing data using t-sne. Journal of machine learning research 9:2579–2605
Selvaraju RR, Cogswell M, Das A, et al (2017) Grad-cam: Visual explanations from deep networks via gradient-based localization. In: ICCV. pp 618–626
Lee H, Yune S, Mansouri M, et al (2018) An explainable deep-learning algorithm for the detection of acute intracranial haemorrhage from small datasets. Nature Biomedical Engineering 1
Chollet F, others (2015) Keras
Abadi M, Barham P, Chen J, et al (2016) Tensorflow: A system for large-scale machine learning. In: OSDI. pp 265–283
Chi J, Walia E, Babyn P, et al (2017) Thyroid nodule classification in ultrasound images by fine-tuning deep convolutional neural network. Journal of digital imaging 30:477–486
Shan J, Alam SK, Garra B, et al (2016) Computer-aided diagnosis for breast ultrasound using computerized bi-rads features and machine learning methods. Ultrasound in medicine & biology 42:980–988
Yeh W-C, Huang S-W, Li P-C (2003) Liver fibrosis grade classification with b-mode ultrasound. Ultrasound in medicine & biology 29:1229–1235
Xian G-m (2010) An identification method of malignant and benign liver tumors from ultrasonography based on glcm texture features and fuzzy svm. Expert Systems with Applications 37:6737–6741
Wu K, Chen X, Ding M (2014) Deep learning based classification of focal liver lesions with contrast-enhanced ultrasound. Optik-International Journal for Light and Electron Optics 125:4057–4063
Acknowledgements
We are sincerely grateful to Dr. Qinghai Peng and Dr. Danming Cao from the Second Xiangya Hospital for their work in expert evaluation.
Funding
This project was funded by RSNA Research Fellow Grant (ID: RF1802) and SIR Foundation Radiology Resident Research Grant to HXB.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study. ILX, HXB and ZZ contributed to conception and design. ILX, JW, JG and HXB contributed to acquisition of data. ILX and HXB contributed to analysis and interpretation of data. ILX, JW, HXB, JG, PJZ, SCH, MCS and ZZ contributed to drafting the article or revising it for important intellectual content. All authors approved the final version to be published.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent for publication
Institutional Review Boards of Hospital of University of Pennsylvania was obtained for the study cohort with waiver of consent.
Ethics approval
The study was approved by the Institutional Review Boards of Hospital of University of Pennsylvania.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
261_2020_2564_MOESM1_ESM.doc
Supplementary file1 Supplementary Fig S1: Confusion matrices for all models and experts. Legend: Confusion matrices of the test datasets for both models are shown. Confusion matricies for both experts are shown as well (DOC 88 kb)
261_2020_2564_MOESM3_ESM.doc
Supplementary file3 Supplementary Table S2: Clinical characteristics of the overall cohort by Code Abdomen categories (DOC 69 kb)
Rights and permissions
About this article
Cite this article
Xi, I.L., Wu, J., Guan, J. et al. Deep learning for differentiation of benign and malignant solid liver lesions on ultrasonography. Abdom Radiol 46, 534–543 (2021). https://doi.org/10.1007/s00261-020-02564-w
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00261-020-02564-w