Abstract
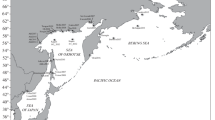
Pacific herring, Clupea pallasii, have recently colonised the northeast Atlantic and Arctic Oceans in the early Holocene. In a relatively short evolutionary time, the herring formed a community with a complex population structure. Previous genetic studies based on morphological, allozyme and mitochondrial DNA data have supported the existence of two herring subspecies from the White Sea and eastern Barents and Kara Seas (C. p. marisalbi and C. p. suworowi, respectively). However, the population structure of the White Sea herring has long been debated and remains controversial. The analyses of morphological and allozyme data have previously identified local spawning groups of herring in the White Sea, whereas mtDNA markers have not revealed any differentiation. We conducted one of the first studies of microsatellite variation for the purpose of investigating the genetic structure and relationship of Pacific herring among ten localities in the White Sea, the Barents Sea and the Kara Sea. Using classical genetic variance-based methods (hierarchical AMOVA, overall and pairwise F ST comparisons), as well as the Bayesian clustering, we infer considerable genetic diversity and population structure in herring at ten microsatellite loci. Genetic differentiation was the most pronounced between the White Sea (C. p. marisalbi) versus the Barents and Kara seas (Chesha–Pechora herring, C. p. suworowi). While microsatellite variation in all C. pallasii was considerable, genetic diversity was significantly lower in C. p. suworowi, than in C. p. marisalbi. Also, tests of genetic differentiation were indicating significant differentiation within the White Sea herring between sympatric summer- and spring-spawning groups, in comparison with genetic homogeneity of the Chesha–Pechora herring.

Similar content being viewed by others
References
ACIA (2004) Impacts of a warming Arctic: Arctic climate impact assessment. ACIA overview report. Cambridge University Press, Cambridge
Altukhov KA (1958) Fishes of the White Sea (in Russian). Nauka, Moscow
Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G (2008) LOSITAN: a workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinforma 9:323
Avise JC (1994) Molecular markers, natural history and evolution. Chapman and Hall, New York
Beacham TD, Schweigert JF, MacConnachie C et al (2002) Population structure of herring (Clupea pallasi) in British Columbia determined by microsatellites, with comparisons to southeast Alaska and California. Can Sci Advis Secr, Res Doc 2002/109. http://www.pac.dfo-mpo.gc.ca/csas
Beerli P (2003) MIGRATE- a maximum likelihood program to estimate gene flow using the coalescent. http://people.scs.fsu.edu/~beerli/download.html
Berger V, Naumov AD (2001) General features. In: Berger V et al (eds) White Sea. Ecology and environment. Derzavets, St. Petersburg, pp 9–20
Buonaccorsi VP, McDowell JR, Graves JE (2001) Reconciling patterns of inter-ocean molecular variance from four classes of molecular markers in blue marlin (Makaira nigricans). Mol Ecol 10:1179–1196
Carvalho GR, Hauser L (1999) Molecular markers and the species concept: New techniques to resolve old disputes? Rev Fish Biol Fish 9:379–382
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Dmitriev NA (1946) Biology and fishery of herring in the White Sea (in Russian). Pishchepromizdat, Moscow
Durand JD, Collet A, Chow S, Guinand B, Borsa P (2005) Nuclear and mitochondrial DNA markers indicate unidirectional gene flow of Indo-Pacific to Atlantic bigeye tuna (Thunnus obesus) populations, and their admixture of southern Africa. Mar Biol 147:313–322
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. doi:10.1007/s12686-011-9548-7
Estoup A, Angers B (1998) Microsatellites and minisatellites for molecular ecology: theoretical and experimental considerations. In: Carvalho GR (ed) Advances in molecular ecology. IOS Press, Amsterdam, pp 55–86
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Evseenko SA, Mishin AV (2011) On the distribution of larvae and localization of spawning stocks of White Sea herring Clupea pallasii marisalbi. J Ichthyol 51:776–787
Evseenko SA, Andrianov DP, Mishin AV, Naumov AP (2006) Species composition and distribution of ichthyoplankton in the White Sea in July 2003. J Ichthyol 46:640–652
Excoffier L, Laval G, Schneider S (2005) ARLEQUIN (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Glover KA, Skaala O, Limborg M, Kvamme C, Torstensen E (2011) Microsatellite DNA reveals population genetic differentiation among sprat (Sprattus sprattus) sampled throughout the north-east Atlantic, including Norwegian fjords. ICES J Mar Sci 68:2145–2151
Goldstein DB, Roemer GW, Smith DA, Reich DE, Bergman A, Wayne RK (1999) The use of microsatellite variation to infer population structure and demographic history in a natural model system. Genetics 151:797–801
Gonzalez EG, Zardoya R (2007) Relative role of life-history traits and historical factors in shaping genetic population structure of sardines (Sardina pilchardus). BMC Evol Biol 7:197. doi:10.1186/1471-2148-7-197
Gonzalez EG, Beerli P, Zardoya R (2008) Genetic structuring and migration patterns of Atlantic bigeye tuna, Thunnus obesus (Lowe, 1839). BMC Evol Biol 8:252. doi:10.1186/1471-2148-8-252
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). http://www.unil.ch/izea/softwares/fstat.html
Grant WS, Bowen BW (1998) Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. J Hered 89:415–426
Guo S, Thompson EA (1992) Performing the exact test of Hardy–Weinberg proportion for multiple alleles. Biometrics 48:361–372
Hardy OJ, Vekemans X (2002) SPAGEDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Hardy OJ, Charbonnel N, Freville H, Heuertz M (2003) Microsatellite allele sizes: a simple test to assess their significance on genetic differentiation. Genetics 163:1467–1482
Hauser L, Carvalho GR (2008) Paradigm shifts in marine fisheries genetics: ugly hypotheses slain by beautiful facts. Fish Fish 9:333–362
Hauser L, Ward RD (1998) Population identification in pelagic fish: the limits of molecular markers. In: Carvalho GR (ed) Advances in molecular ecology. IOS Press, Amsterdam, pp 191–224
Hauser L, Adcock GJ, Smith PJ et al (2002) Loss of microsatellite diversity and low effective population size in an overexploited population of New Zealand snapper (Pagrus auratus). Proc Natl Acad Sci USA 99:11742–11747
Hedgecock D (1994) Does variance in reproductive success limit effective population size of marine organisms? In: Beaumont A (ed) Genetics and evolution of aquatic organisms. Chapman and Hall, London, pp 122–134
Hedrick PW (1986) Genetic polymorphism in heterogeneous environments: a decade later. Annu Rev Ecol Syst 17:535–566
Hewitt GM (1996) Some genetic consequences of ice ages and their role in divergence and speciation. Biol J Linn Soc 58:247–276
Hutchinson DW, Templeton AR (1999) Correlation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution 53:1898–1914
Jensen JL, Bohonak AJ, Kelley ST (2005) Isolation by distance, web service. BMC Genet 6:13. http://ibdws.sdsu.edu/
Johannesson K, André C (2006) Life on the margin: genetic isolation and diversity loss in a peripheral marine ecosystem, the Baltic Sea. Mol Ecol 15:2013–2029
Jørstad KE, Novikov GG, Stasenkova NJ et al (2001) Intermingling of herring stocks in the Barents Sea area. In: Balckburn FF et al (eds) Herring: expectations for a new millennium. Fairbanks, Alaska, pp 629–633
Koizumi I, Yamamoto S, Maekawa K (2006) Decomposed pairwise regression analysis of genetic and geographic distances reveals a metapopulation structure of stream-dwelling Dolly Varden charr. Mol Ecol 15:3175–3189
Kulakov MY, Pogrebov VB, Timofeyev SF, Chernova NV, Kiyko OA (2006) Ecosystems of the Barents and Kara seas, coastal segment. In: Robinson AR, Brin KH (eds) The sea ideas and observations on progress in the study of sea, V 14. Harvard University Press, Cambridge, pp 1135–1172
Laakkonen HM, Lajus DM, Strelkov P, Väinölä R (2013) Phylogeography of amphi-boreal fish: tracing the history of the Pacific herring Clupea pallasii in North-East European seas. BMC Evol Biol 13:67. doi:10.1186/1471-2148-13-67
Laikre L, Palm S, Ryman N (2005) Genetic population structure of fishes: implications for coastal zone management. Ambio 34:111–119
Lajus DL (1996) White Sea herring (Clupea pallasi marisalbi, Berg) population structure: interpopulation variation in frequency of chromosomal rearrangement. Cybium 20(3):279–294
Lajus DL (2002) Long-term discussion on the stocks of the White Sea herring: historical perspective and present state. ICES Mar Sci Symp 215:321–328
Larsson LC, Laikre L, André C, Dahlgren TG, Ryman N (2010) Temporally stable genetic structure of heavily exploited Atlantic herring (Clupea harengus) in Swedish waters. Heredity 104:40–51
Lesica P, Allendorf FW (1995) When are peripheral populations valuable for conservation? Conserv Biol 9:753–760
Lewis PO, Zaykin D (2001) Genetic data analysis: Computer program for the analysis of allelic data: version 1.0 (d 16c). http://lewis.eeb.unconn.edu/lewishome/software.html
Limborg MT, Pedersen JS, Hemmer-Hansen J, Tomkiewicz J, Bekkevold D (2009) Genetic population structure of European sprat, Sprattus sprattus: differentiation across a steep environmental gradient in a small pelagic fish. Mar Ecol Prog Ser 379:213–224
Limborg MT, Helyar SJ, de Bruyn M et al (2012) Environmental selection on transcriptome-derived SNPs in a high gene flow marine fish, the Atlantic herring (Clupea harengus). Mol Ecol 21:3686–3703
Liu M, Lin L, Gao T, Yanagimoto T, Sakurai Y et al (2012) What maintains the central North Pacific genetic discontinuity in Pacific herring? PLoS One 7(12):e50340. doi:10.1371/journal.pone.0050340
Manni F, Guérard E, Heyer E (2004) Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by “Monmonier’s algorithm”. Hum Biol 76(2):173–190
Matishov G, Zuyev A, Golubev V et al (2004) Climatic atlas of the Arctic seas 2004: part I. Database of the Barents, Kara, Laptev, and White Seas-oceanography and marine biology NOAA Atlas NESDIS 58. U.S. Gov. Printing Office, Washington DC
McPherson AA, O’Reilly PT, McParland TL et al (2001) Isolation of nine novel tetranucleotide microsatellites in Atlantic herring (Clupea harengus). Mol Ecol Notes 1:31–32
Miller KM, Laberee K, Schulze AD, Kaukinen KH (2001) Development of microsatellite loci in Pacific herring (Clupea pallasi). Mol Ecol Notes 1:131–132
Mills LS, Allendorf FW (1996) The one-migrant-per-generation rule in conservation and management. Conserv Biol 6:1509–1518
Mishin AV, Evseenko SA, Evdokimov YuV (2008) Species composition and distribution of summer ichthyoplankton in Chupa Estuary (Kandalaksha Bay of the White Sea). J Ichthyol 48:80–816
O’Connell M, Dillon MC, Wright JM (1998) Development of primers for polymorphic microsatellite loci in the pacific herring (Clupea harengus pallasi). Mol Ecol 7:358–360
Olafsson K, Pampoulie C, Hjorleifsdottir S, Gudjonsson S, Hreggvidsson GO (2014) Present-day genetic structure of Atlantic Salmon (Salmo salar) in Icelandic rivers and Ice-Cap retreat models. PLoS One 9(2):e86809
Olsen JB, Lewis CJ, Kretschmer EJ et al (2002) Characterization of 14 tetranucleotide microsatellite loci derived from Pacific herring. Mol Ecol Notes 2:101–103
Palstra FP, Fraser DJ (2012) Effective/census population size ratio estimation: a compendium and appraisal. Ecol Evol 2(9):2357–2365
Pantyulin AN (2003) Hydrological system of the White Sea. Oceanology 43:1–14
Peery MZ, Kirby R, Reid BN et al (2012) Reliability of genetic bottleneck tests for detecting recent population declines. Mol Ecol 21:3403–3418
Piry S, Luikart G, Conuet JM (1999) Bottleneck: a computer program for detecting recent reduction in the effective population size using allele frequency data. J Hered 90:502–503
Pritchard J, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86(5):248–249
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Ruzzante DE, Mariani S, Bekkevold D et al (2006) Biocomplexity in a highly migratory pelagic marine fish, Atlantic herring. Proc R Soc B 273:1459–1464. doi:10.1098/rspb.2005.3463
Ryman N, Palm S, André C, Carvalho GR, Dahlgren TG et al (2006) Power for detecting genetic divergence: differences between statistical methods and marker loci. Mol Ecol 15:2031–2045
Schwartz MK, Mills LS, Ortega Y, Ruggiero LF, Allendorf FW (2003) Landscape location affects genetic variation of Canada lynx (Lynx canadensis). Mol Ecol 12:1807–1816
Semenova AV, Andreeva AP, Karpov AK, Frolov SB, Feoktistov EI, Novikov GG (2004) Genetic variation of herrings of the genus Clupea from the White Sea. J Ichthyol 44:229–238
Semenova AV, Andreeva AP, Karpov AK, Novikov GG (2009) An analysis of allozyme variation in herring Clupea pallasii from the White and Barents seas. J Ichthyol 49:313–330. doi:10.1134/S0032945209040043
Semenova AV, Andreeva AP, Karpov AK, Stroganov AN, Rubtsova GA, Afanasiev KI (2013) Analysis of microsatellite loci variations in herring (Clupea pallasii marisalbi) from the White Sea. Russ J Genet 49:652–666. doi:10.1134/S1022795413060100
Semenova AV, Stroganov AN, Smirnov AA, Afanasiev KI, Rubtsova GA (2014) Genetic variation of herring Clupea pallasii from the Sea of Okhotsk revealed by microsatellite. Russ J Genet 50:65–69. doi:10.1134/S1022795413120107
Shikano T, Ramadevi J, Merilä J (2010) Identification of local- and habitat-dependent selection: scanning functionally important genes in nine-spined sticklebacks (Pungitius pungitius). Mol Biol Evol 27(12):2775–2789
Shrimpton JM, Health DD (2003) Census vs. effective population size in chinook salmon: large- and small-scale environmental perturbation effects. Mol Ecol 10:2571–2583
Small MP, Loxterman JL, Frye AE, Von Bargen JF, Bowman C, Young SF (2005) Temporal and spatial genetic structure among some Pacific herring (Clupea pallasi) populations in Puget Sound and the Southern Strait of Georgia. Trans Am Fish Soc 134:1329–1341. doi:10.1577/T05-050.1
Sugaya T, Sato M, Yokoyama E et al (2008) Population genetic structure and variability of Pacific herring Clupea pallasii in the stocking area along the Pacific coast of northern Japan. Fish Sci 74:579–588. doi:10.1111/j.1444-2906.2008.01561.x
Sunnuks P (2000) Efficient genetic markers for population biology. Trends Ecol Evol 15:199–203
Svetovidov AN (1952) Clupeidae. In: Fauna SSSR. The fishes 2(1) (In Russian). Zoologicheskii Institut Akademiya Nauk SSSR, Moscow
Teacher AGF, Kähkönen K, Merilä J (2011) Development of 61 new transcriptome-derived microsatellites for the Atlantic herring (Clupea harengus). Conserv Genet Resour. doi:10.1007/s12686-011-9477-5
Teacher AGF, André C, Jonsson PR, Merilä J (2013) Oceanographic connectivity and environmental correlates of genetic structuring in Atlantic herring in the Baltic Sea. Evol Appl. doi:10.1111/eva.12042
Vähä J-P, Erkinaro J, Niemelä E, Primmer CR (2007) Life-history and habitat features influence the within-river genetic structure of Atlantic salmon. Mol Ecol 16:2638–2654
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Ward RD, Woodmark M, Skibinski DOF (1994) A comparison of genetic diversity levels in marine, freshwater and anadromous fishes. J Fish Biol 44:213–232
Warnock WG, Rasmussen JB, Taylor EB (2010) Genetic clustering methods reveal bull trout (Salvelinus confluentus) fine-scale population structure as a spatially nested hierarchy. Conserv Genet 11:1421–1433
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Whittaker JC, Harbord RM, Boxall N et al (2003) Likelihood-based estimation of microsatellite mutation rates. Genetics 164(2):781–787
Wildes SL, Vollenweider JJ, Nguyen HT, Guyon JR (2011) Genetic variation between outer-coastal and fjord populations of Pacific herring (Clupea pallasii) in the eastern Gulf of Alaska. Fish Bull 109:382–393
Acknowledgments
This work was supported by the Russian Foundation for Basic Research (Grant No. 13-04-00247-a) and the Leading Scientific Schools Program (Project NSh2666.2014.4).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Semenova, A.V., Stroganov, A.N., Afanasiev, K.I. et al. Population structure and variability of Pacific herring (Clupea pallasii) in the White Sea, Barents and Kara Seas revealed by microsatellite DNA analyses. Polar Biol 38, 951–965 (2015). https://doi.org/10.1007/s00300-015-1653-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00300-015-1653-8