Abstract
Key message
An SVP protein, PhSVP, bound to the CArG-boxes in the promoter regions of FT-like paralogs and repressed their expression, thus affecting the floral transition in Phalaenopsis orchid.
Abstract
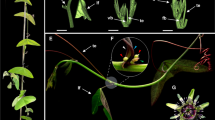
Phalaenopsis is an important ornamental flower native to tropical rain forests. It usually reaches vegetative maturity after 4–5 leaves and, after a juvenile stage, forms a flower spike (inflorescence) from the axillary buds. The PEBP gene family encodes a phosphatidyl-ethanolamine-binding protein (PEBP) domain involved in regulating flowering and other aspects of plant development. Here, we identified eight PEBP family genes in Phalaenopsis and detected the expression patterns of seven of them in various organs. Among them, PhFT1 (Phalaenopsis hybrid FLOWERING LOCUS T1), PhFT3, PhFT5, and PhMFT (Phalaenopsis hybrid MOTHER OF FT AND TFL1) promoted flowering in transgenic Arabidopsis, while PhFT6 inhibited flowering. PhSVP (Phalaenopsis hybrid SHORT VEGETATIVE PHASE), an SVP protein that repressed flowering in Arabidopsis, bound to the CArG-boxes in the promoter regions of PhFT3, PhFT6, and PhMFT in a yeast one-hybrid assay. Additionally, dual-luciferase and transient expression assays showed that PhSVP significantly inhibits the expression of both PhFT3 and PhFT6. Together, our work provides a comprehensive understanding of the PhFT-like genes that can promote or repress flowering, and it suggests strategies for regulating the floral transition in Phalaenopsis that exploit the evolutionary versatility of PhFTs to respond to various signals stimuli.










Similar content being viewed by others
References
Araki T, Kobayashi Y, Kaya H, Iwabuchi M (1998) The flowering-time gene FT and regulation of flowering in Arabidopsis. J Plant Res 111:277–281
Blackman BK, Strasburg JL, Raduski AR, Michaels SD, Rieseberg LH (2010) The role of recently derived FT paralogs in sunflower domestication. Curr Biol 20:629–635
Blanchard MG, Runkle ES (2006) Temperature during the day, but not during the night, controls flowering of Phalaenopsis orchids. J Exp Bot 57:4043–4049
Capovilla G, Schmid M, Posé D (2015) Control of flowering by ambient temperature. J Exp Bot 66:59–69
Chardon F, Damerval C (2005) Phylogenomic analysis of the PEBP gene family in cereals. J Mol Evol 61:579–590
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Coelho CP, Minow MA, Chalfun-Junior A, Colasanti J (2014) Putative sugarcane FT/TFL1 genes delay flowering time and alter reproductive architecture in Arabidopsis. Front Plant Sci 5:221
Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316:1030–1033
Danilevskaya ON, Meng X, Hou Z, Ananiev EV, Simmons CR (2008) A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol 146:250–264
Fu C, Chen Y, Hsiao Y, Pan Z, Liu Z, Huang Y, Tsai W, Chen H (2011) OrchidBase: a collection of sequences of the transcriptome derived from orchids. Plant Cell Physiol 52:238–243
Goh CJ, Yang AL (1978) Effects of growth regulators and decapitation on flowering of Dendrobium orchid hybrids. Plant Sci Lett 12:287–292
Harig L, Beinecke FA, Oltmanns J, Muth J, Mueller O, Rueping B, Twyman RM, Fischer R, Pruefer D, Noll GA (2012) Proteins from the FLOWERING LOCUS T-like subclade of the PEBP family act antagonistically to regulate floral initiation in tobacco. Plant J 72:908–921
Higuchi Y (2018) Florigen and anti-florigen: flowering regulation in horticultural crops. Breed Sci 68:109–118
Ho WWH, Weigel D (2014) Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell 26:552–564
Hou C, Yang C (2009) Functional analysis of FT and TFL1 orthologs from orchid (Oncidium Gower Ramsey) that regulate the vegetative to reproductive transition. Plant Cell Physiol 50:1544–1557
Huang W, Fang Z, Zeng S, Zhang J, Wu K, Chen Z, Teixeira da Silva JA, Duan J (2012) Molecular cloning and functional analysis of three FLOWERING LOCUS T (FT) homologous genes from Chinese Cymbidium. Int J Mol Sci 13:11385–11398
Jang S, Choi S, Li H, An G, Schmelzer E (2015) Functional characterization of Phalaenopsis aphrodite flowering genes PaFT1 and PaFD. PLoS ONE. https://doi.org/10.1371/journal.pone.0134987
Karlgren A, Gyllenstrand N, Kallman T, Sundstrom JF, Moore D, Lascoux M, Lagercrantz U (2011) Evolution of the PEBP gene family in plants: functional diversification in seed plant evolution. Plant Physiol 156:1967–1977
Kikuchi R, Kawahigashi H, Ando T, Tonooka T, Handa H (2009) Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiol 149:1341–1353
Koornneef M, Hanhart CJ, Vanderveen JH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229:57–66
Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH (2007) Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev 21:397–402
Lee R, Baldwin S, Kenel F, McCallum J, Macknight R (2013) FLOWERING LOCUS T genes control onion bulb formation and flowering. Nat Commun 4:2884
Li D, Liu C, Shen L, Wu Y, Chen H, Robertson M, Helliwell CA, Ito T, Meyerowitz E, Yu H (2008) A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell 15:110–120
Li R, Wang A, Sun S, Liang S, Wang X, Ye Q, Li H (2012) Functional characterization of FT and MFT ortholog genes in orchid (Dendrobium nobile Lindl) that regulate the vegetative to reproductive transition in Arabidopsis. Plant Cell Tissue Organ Cult 111:143–151
Li J, Liao X, Zhou S, Liu S, Jiang L, Wang G (2018) Efficient protoplast isolation and transient gene expression system for Phalaenopsis hybrid cultivar ‘Ruili Beauty.’ In Vitro Cell Dev Biol-Plant 54:87–93
Liu X, Ning K, Che G, Yan S, Han L, Gu R, Li Z, Weng Y, Zhang X (2018) CsSPL functions as an adaptor between HD-ZIPIII and CsWUS transcription factors regulating anther and ovule development in Cucumis sativus (cucumber). Plant J 94:535–547
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408
Lopez RG, Runkle ES (2005) Environmental physiology of growth and flowering of orchids. HortScience 40:1969–1973
Ohshima S, Murata M, Sakamoto W, Ogura Y, Motoyoshi F (1997) Cloning and molecular analysis of the Arabidopsis gene Terminal Flower 1. Mol Gen Genet 254:186–194
Ospina-Zapata DA, Madrigal Y, Alzate JF, Pabón-Mora N (2020) Evolution and expression of reproductive transition regulatory genes FT/TFL1 with emphasis in selected neotropical orchids. Front Plant Sci 11:469
Pin PA, Benlloch R, Bonnet D, Wremerth-Weich E, Kraft T, Gielen JJL, Nilsson O (2010) An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330:1397–1400
Posé D, Yant L, Schmid M (2012) The end of innocence: flowering networks explode in complexity. Curr Opin Plant Biol 15:45–50
Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc-finger transcription factors. Cell 80:847–857
Rombauts S, Dehais P, Van Montagu M, Rouze P (1999) PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res 27:295–296
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shannon S, Meeks-Wagner DR (1991) A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. Plant Cell 3:877–892
Shu K, Luo X, Meng Y, Yang W (2018) Toward a molecular understanding of abscisic acid actions in floral transition. Plant Cell Physiol 59:215–221
Tang X, Liang M, Han J, Cheng J, Zhang H, Liu X (2020) Ectopic expression of LoSVP, a MADS-domain transcription factor from lily, leads to delayed flowering in transgenic Arabidopsis. Plant Cell Rep 39:289–298
Teo ZWN, Zhou W, Shen L (2019) Dissecting the function of MADS-Box transcription factors in orchid reproductive development. Front Plant Sci 10:1474
Teper-Bamnolker P, Samach A (2005) The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17:2661–2675
Tocquin P, Corbesier L, Havelange A, Pieltain A, Kurtem E, Bernier G, Perilleux C (2003) A novel high efficiency, low maintenance, hydroponic system for synchronous growth and flowering of Arabidopsis thaliana. BMC Plant Biol 3:2
Tsuji H (2017) Molecular function of florigen. Breed Sci 67:327–332
Wang H, Tong C, Jang S (2017a) Current progress in orchid flowering/flower development research. Plant Signal Behav. https://doi.org/10.1080/15592324.2017.1322245
Wang Y, Liu L, Song S, Li Y, Shen L, Yu H (2017b) DOFT and DOFTIP1 affect reproductive development in the orchid Dendrobium Chao Praya smile. J Exp Bot 8:5759–5772
Wang Z, Wang F, Hong Y, Yao J, Ren Z, Shi H, Zhu JK (2018) The flowering repressor SVP confers drought resistance in Arabidopsis by regulating abscisic acid catabolism. Mol Plant 11:1184–1197
Wang S, Viswanath KK, Tong C, An HR, Jang S, Chen F (2019) Floral induction and flower development of orchids. Front Plant Sci 10:1258
Wickland DP, Hanzawa Y (2015) The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: functional evolution and molecular mechanisms. Mol Plant 8:983–997
Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309:1056–1059
William C (2004) Understanding orchids, 1st edn. Houghton Mifflin Company, New York
Winterhagen P, Tiyayon P, Samach A, Hegele M, Wunsche JN (2013) Isolation and characterization of FLOWERING LOCUS T subforms and APETALA1 of the subtropical fruit tree Dimocarpus longan. Plant Physiol Bioch 71:184–190
Xiang L, Li X, Qin D, Guo F, Wu C, Miao L, Sun C (2012) Functional analysis of FLOWERING LOCUS T orthologs from spring orchid (Cymbidium goeringii Rchb. f.) that regulates the vegetative to reproductive transition. Plant Physiol Bioch 58:98–105
Yang F, Zhu G, Wei Y, Gao J, Liang G, Peng L, Lu C, Jin J (2019) Low-temperature-induced changes in the transcriptome reveal a major role of CgSVP genes in regulating flowering of Cymbidium goeringii. BMC Genomics 20:53
Yoo SY, Kardailsky I, Lee JS, Weigel D, Ahn JH (2004) Acceleration of flowering by overexpression of MFT (MOTHER OF FT AND TFL1). Mol Cells 17:95–101
Zhai H, Lu S, Liang S, Wu H, Zhang X, Liu B, Kong F, Yuan X, Li J, Xia Z (2014) GmFT4, a homolog of FLOWERING LOCUS T, is positively regulated by E1 and functions as a flowering repressor in soybean. PLoS ONE. https://doi.org/10.1371/journal.pone.0089030
Zhang J, Yan S, Zhao W, Zhang X (2013) Cloning and functional analysis of CsFT in cucumber. Acta Hortic Sinica 40:2180–2188 (In Chinese)
Zhao W, Gu R, Che G, Cheng Z, Zhang X (2018) CsTFL1b may regulate the flowering time and inflorescence architecture in cucumber (Cucumis sativus L.). Biochem Biophys Res Commun 499:307–313
Zheng X, Wu F, Zhang X, Lin Q, Wang J, Guo X, Lei C, Cheng Z, Zou C, Wan J (2016) Evolution of the PEBP gene family and selective signature on FT-like clade. J Syst Evol 54:502–510
Zhou S, Jiang L, Guan S, Gao Y, Gao Q, Wang G, Duan K (2018) Expression profiles of five FT-like genes and functional analysis of PhFT-1 in a Phalaenopsis hybrid. Electron J Biotechn 31:75–83
Acknowledgements
The authors would like to thank Prof. Xiaolan Zhang of China Agricultural University for sharing the ft-10 and tfl1-11 seeds and vectors for the yeast assays, Dr. Ze Wu of Nanjing Agricultural University for providing vectors for the dual-luciferase reporter assay, and Dr. Ji Li of Nanjing Agricultural University for proving the pGreen vector. This work was supported by the National Natural Science Foundation of China (No. 31372101 and No. 31701960) and the National Key R&D Program of China (2020YFD1000400).
Author information
Authors and Affiliations
Contributions
GW and LJ designed the experiments, analyzed the data and wrote the paper. LJ, XJ, YL, YG, SW and YM performed the experiments.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Da-Bing Zhang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

299_2021_2805_MOESM3_ESM.jpg
Supplementary file3 (JPG 2280 KB) Cis-regulatory element analysis of PhFT3, PhFT6 and PhMFT and their homologs from Arabidopsis. The promoter regions (2–3 kb upstream of the transcription start site) were scanned for cis-regulatory elements. Eight groups of cis-regulatory elements (abiotic, biotic, tissue-specific, light responsive, core promoter element, auxin responsive, cell cycle and others) are listed on the x axis, and gene names are listed on the y axis. The color key from white (low) to dark orange (high) indicates the number of cis-regulatory elements in the promoter region of each gene.

299_2021_2805_MOESM4_ESM.jpg
Supplementary file4 (JPG 1337 KB) Phylogenetic tree of PhSVP and other plant SVP proteins. The red dot marks the SVP from Phalaenopsis. At, Arabidopsis thaliana; Cg, Cymbidium goeringii; As, Apostasia shenzhenica; Dc, Dendrobium catenatum; Lp, Lilium pumilum; Nn, Nelumbo nucifera; Vv, Vitis vinifera; Ac, Actinidia chinensis. The accession numbers or gene ID numbers of SVPs in TAIR, GenBank or OrchidBase are as follows: PhSVP (MG560826), AtSVP1 (AT2G22540), AtSVP2 (AT4G24540), CgSVP1 (MF462083), CgSVP2 (MF462098), CgSVP3 (MF462082), AsSVP (Ash016779), DcSVP (Dca020215), LpSVP (AXE75656), NnSVP (XP_010254525), VvSVP (XP_019073897), AcSVP1 (JF838216), AcSVP2 (JF838217), AcSVP3 (JF838218) and AcSVP4 (JF838219).

299_2021_2805_MOESM5_ESM.jpg
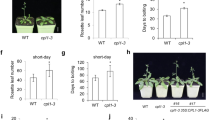
Supplementary file5 (JPG 795 KB) Heterologous expression of PhSVP in wild-type Arabidopsis. (a) Phenotypes of Pro35S:PhSVP#1 and Pro35S:PhSVP#2 transgenic plants. (b) Days to flowering and rosette leaf number of Pro35S:PhSVP transgenic plants. V/C, empty-vector/control Arabidopsis (Pro35S:GFP/Col-0). Asterisks represent significant differences derived from one-way ANOVA followed by Dunnett‘s multiple comparisons tests (**, P < 0.01).

299_2021_2805_MOESM6_ESM.jpg
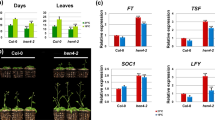
Supplementary file6 (JPG 1214 KB) Phenotypic characterizations of Pro35S:PhFT1/ft-10 and Pro35S:PhFT5/ft-10 transgenic plants.
Rights and permissions
About this article
Cite this article
Jiang, L., Jiang, X., Li, Y. et al. FT-like paralogs are repressed by an SVP protein during the floral transition in Phalaenopsis orchid. Plant Cell Rep 41, 233–248 (2022). https://doi.org/10.1007/s00299-021-02805-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-021-02805-2