Abstract
Key message
WsWRKY1-mediated transcriptional modulation of Withania somnifera tryptophan decarboxylase gene (WsTDC) helps to regulate fruit-specific tryptamine generation for production of withanamides.
Abstract
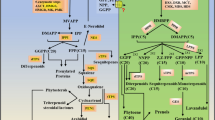
Withania somnifera is a highly valued medicinal plant. Recent demonstration of novel indolyl metabolites called withanamides in its fruits (berries) prompted us to investigate its tryptophan decarboxylase (TDC), as tryptophan is invariably a precursor for indole moiety. TDC catalyzes conversion of tryptophan into tryptamine, and the catalytic reaction constitutes a committed metabolic step for synthesis of an array of indolyl metabolites. The TDC gene (WsTDC) was cloned from berries of the plant and expressed in E. coli. The recombinant enzyme was purified and characterized for its catalytic attributes. Catalytic and structural aspects of the enzyme indicated its regulatory/rate-limiting significance in generation of the indolyl metabolites. Novel tissue-wise and developmentally differential abundance of WsTDC transcripts reflected its preeminent role in withanamide biogenesis in the fruits. Transgenic lines overexpressing WsTDC gene showed accumulation of tryptamine at significantly higher levels, while lines silenced for WsTDC exhibited considerably depleted levels of tryptamine. Cloning and sequence analysis of promoter of WsTDC revealed the presence of W-box in it. Follow-up studies on isolation of WsWRKY1 transcription factor and its overexpression in W. somnifera revealed that WsTDC expression was substantially induced by WsWRKY1 resulting in overproduction of tryptamine. The study invokes a key role of TDC in regulating the indolyl secondary metabolites through enabling elevated flux/supply of tryptamine at multiple levels from gene expression to catalytic attributes overall coordinated by WsWRKY1. This is the first biochemical, molecular, structural, physiological and regulatory description of a fruit-functional TDC.











Similar content being viewed by others
References
Ahmad H, Arya A, Agrawal S, Samuel SS, Singh SK, Valicherla GR, Sangwan N, Mitra K, Gayen JR, Paliwal S, Shukla R, Dwivedi AK (2016) Phospholipid complexation of NMITLI118RT+: way to a prudent therapeutic approach for beneficial outcomes in ischemic stroke in rats. Drug Deliv. 23(9):3606–3618
Akula R, Giridhar P, Ravishankar GA (2011) Phytoserotonin: a review. Plant Signal Behav 6:800–809
Badria FA (2002) Melatonin, serotonin and tryptamine in some Egyptian food and medicinal plants. J Med Food 5:153–157
Burkhard P, Dominici P, Borri-Voltattorni C, Jansonius JN, Malashkevich VN (2001) Structural insight into Parkinson’s disease treatment from drug-inhibited DOPA decarboxylase. Nat Struct Biol 8:963–967
Chaurasiya ND, Sangwan RS, Misra LN, Tuli R, Sangwan NS (2009) Metabolic clustering of a core collection of Indian ginseng (Withania somnifera) through DNA, isoenzymes, polypeptide and withanolide profile diversity. Fitoterapia 80:496–505
Chen H, Lai Z, Shi J, Xiao Y, Chen Z, Xu X (2010) Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol 10:281
Di Fiore S, Li Q, Leech MJ, Schuster F, Emans N, Fischer R, Schillberg S (2002) Targeting tryptophan decarboxylase to selected subcellular compartments of tobacco plants affects enzyme stability and in vivo function and leads to a lesion-mimic phenotype. Plant Physiol 129:1160–1169
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5:199–206
Facchini PJ, Huber-Allanach KL, Tari LW (2000) Plant aromatic l-amino acid decarboxylases: evolution, biochemistry, regulation, and metabolic engineering applications. Phytochemistry 54:121–138
Gibson RA, Barret G, Wightman F (1972) Biosynthesis and metabolism of indole-3-acetic acid: partial purification and properties of a tryptamine-forming l-tryptophan decarboxylase from tomato shoots. J Exp Bot 23:775–786
Gupta P, Akhtar N, Tewari SK, Sangwan RS, Trivedi PK (2011) Differential expression of farnesyl diphosphate synthase gene from Withania somnifera in different chemotypes and in response to elicitors. Plant Growth Regul 65:93–100
Ishihara A, Nakao T, Mashimo Y, Murai M, Ichimaru N, Tanaka C, Nakajima H, Wakasa K, Miyagawa H (2011) Probing the role of tryptophan-derived secondary metabolism in defense responses against Bipolaris oryzae infection in rice leaves by a suicide substrate of tryptophan decarboxylase. Phytochemistry 72:7–13
Jadaun JS, Sangwan NS, Narnoliya LK, Tripathi S, Sangwan RS (2016) Withania coagulans tryptophan decarboxylase gene cloning, heterologous expression and catalytic characteristics of the recombinant enzyme. Protoplasma 254(1):181–192
Jadaun JS, Sangwan NS, Narnoliya LK, Singh N, Bansal S, Mishra B, Sangwan RS (2017) Over-expression of DXS gene enhances terpenoidal secondary metabolite accumulation in rose-scented geranium and Withania somnifera: active involvement of plastid isoprenogenic pathway in their biosynthesis. Physiol Plant 159(4):381–400
Jayaprakasham B, Padmanabhan K, Nair MG (2010) Withanamides in Withania somnifera fruit protect PC-12 cells from β-amyloid responsible for Alzheimer’s disease. Phytother Res 24:859–863
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901
Jiang Y, Duan Y, Yin J, Ye S, Zhu J, Zhang F, Lu W, Fan D, Luo K (2014) Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J Exp Bot 65(22):6629–6644
Kang S, Kang K, Lee K, Back K (2007) Characterization of rice tryptophan decarboxylases and their direct involvement in serotonin biosynthesis in transgenic rice. Planta 227:263–272
Kang K, Kang S, Lee K, Park M (2008) Enzymatic features of serotonin biosynthetic enzyme and serotonin biosynthesis in plants. Plant Signal Behav 3:389–390
Kato N, Kojima T, Yoshiyagawa S, Ohta T, Toriba A, Nishimura H, Hayakawa K (2007) Rapid and sensitive determination of tryptophan, serotonin and psychoactive tryptamines by thin-layer chromatography/fluorescence detection. J Chromatogr A 1145:229–233
Khedgikar V, Ahmad N, Kushwaha P, Gautam J, Nagar GK, Singh D, Trivedi PK, Mishra PR, Sangwan NS, Trivedi R (2015) Preventive effects of withaferin A isolated from the leaves of an Indian medicinal plant Withania somnifera (L.): Comparisons with 17-β-estradiol and alendronate. Nutrition 31(1):205–213
Kushwaha AK, Sangwan NS, Trivedi PK, Negi AS, Misra L, Sangwan RS (2013) Tropine forming tropinone reductase gene from Withania somnifera (Ashwagandha): biochemical characteristics of the recombinant enzyme and novel physiological overtones of tissue-wide gene expression patterns. PLoS ONE 8:e74777
Leon J, Rojo E, Sanchez J (2001) Wound signalling in plants. J Exp Bot 52:1–9
Liu W, Chen R, Chen M, Zhang H, Peng M, Yang C, Ming X, Lan X, Liao Z (2012) Tryptophan decarboxylase plays an important role in ajmalicine biosynthesis in Rauvolfia verticillata. Planta 236:239–250
López-Meyse M, Nessler CL (1997) Tryptophan decarboxylase is encoded by two autonomously regulated genes in Camptotheca acuminata which are differentially expressed during development and stress. Plant J 11:1167–1175
Mishra S, Bansal S, Sangwan RS, Sangwan NS (2016) Genotype independent and efficient Agrobacterium-mediated genetic transformation of the medicinal plant Withania somnifera Dunal. J Plant Biochem Biot 25(2):191–198
Mishra B, Bose SK, Sangwan NS (2020) Comparative investigation of therapeutic plant Withania somnifera for yield, productivity, withanolide contents, and expression of pathway genes during contrasting seasons. Ind Crops Prod 154:112508
Noe W, Mollenschott C, Berlin J (1984) Tryptophan decarboxylase from Catharanthus roseus cell suspension cultures: purification, molecular and kinetic data of the homogenous protein. Plant Mol Biol 3:281–288
Ouwerkerk PBF, Memelink J (1999) Elicitor-responsive promoter regions in the tryptophan decarboxylase gene from Catharanthus roseus. Plant Mol Biol 39:129–213
Rushton DL, Tripathi P, Rabara RC, Lin J, Ringler P, Boken AK, Langum TJ, Smidt L, Boomsma DD, Emme NJ, Chen X (2012) WRKY transcription factors: key components in abscisic acid signalling. Plant Biotech J 10:2–11
Sabir F, Mishra S, Sangwan RS, Jadaun JS, Sangwan NS (2013) Qualitative and quantitative variations in withanolides and expression of some pathway genes during different stages of morphogenesis in Withania somnifera Dunal. Protoplasma 250:539–549
Sangwan RS, Mishra S, Kumar S (1998) Direct fluorometry of phase-extracted tryptamine-based fast quantitative assay of l-tryptophan decarboxylase from Catharanthus roseus leaf. Anal Biochem 255:39–46
Sangwan RS, Chaurasiya ND, Misra LN, Lal P, Uniyal GC, Sangwan NS, Srivastava AK, Suri KA, Qazi GN and Tuli R (2005) An improved process for isolation of withaferin-A from plant materials and products therefrom. US Patent 7,108, 870
Sangwan RS, Chaurasiya ND, Lal P, Misra L, Tuli R, Sangwan NS (2008) Root-contained withanolide A is inherently de novo synthesized within roots in Ashwagandha (Withania somnifera). Physiol Plant 133:278–287
Sangwan NS, Sangwan RS (2014) Secondary metabolites of traditional medical plants: a case study of Ashwagandha (Withania somnifera). In: Nick P, Opatrrny Z (eds) Applied Plant Cell Biology, Plant cell monograph. Springer, Verlag, Berlin Heidelberg, pp 325–367
Schluttenhofer C, Pattanaik S, Patra B, Yuan L (2014) Analyses of Catharanthus roseus and Arabidopsis thaliana WRKY transcription factors reveal involvement in jasmonate signaling. BMC Genomics 15:1
Sharma PK, Sangwan NS, Bose SK, Sangwan RS (2013) Biochemical characteristics of a novel vegetative tissue geraniol acetyltransferase from a monoterpene oil grass (Palmarosa, Cymbopogon martinii var. Motia) leaf. Plant Sci 203:63–73
Sheen J (2001) Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol 127:1466–1475
Songstad DD, De Luca V, Brisson N, Kurz WG, Nessler CL (1990) High levels of tryptamine accumulation in transgenic tobacco expressing tryptophan decarboxylase. Plant Physiol 94:1410–1413
Srivastava S, Sangwan RS, Tripathi S, Mishra B, Narnoliya LK, Misra LN, Sangwan NS (2015) Light and auxin responsive cytochrome P450s from Withania somnifera Dunal: cloning, expression and molecular modelling of two pairs of homologue genes with differential regulation. Protoplasma 252:1421–1437
Stevens LH, Blom TJM, Verpoorte R (1993) Subcellular localization of tryptophan decarboxylase, strictosidine synthase and strictosidine glucosidase in suspension cultured cells of Catharanthus roseus and Tabernaemontana divaricata. Plant Cell Rep 12:573–576
Sun Y, Niu Y, Xu J, Li Y, Luo H, Zhu Y, Liu M, Wu Q, Song J, Sun C, Chen S (2013) Discovery of WRKY transcription factors through transcriptome analysis and characterization of a novel methyl jasmonate-inducible PqWRKY1 gene from Panax quinquefolius. Plant Cell Tiss Org 114:269–277
Suttipanta N, Pattanaik S, Kulshrestha M, Patra B, Sing SK, Yuan L (2011) The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol 157:2081–2093
Torrens-Spence MP, Lazear M, Von Guggenberg R, Ding H, Li J (2014) Investigation of a substrate-specifying residue within Papaver somniferum and Catharanthus roseus aromatic amino acid decarboxylases. Phytochemistry 106:37–43
Tripathi S, Sangwan RS, Narnoliya LK, Srivastava Y, Mishra B, Sangwan Neelam S (2017) Transcription factor repertoire in Ashwagandha (Withania somnifera) through analytics of transcriptomic resources: Insights into regulation of development and withanolide metabolism. Sci Rep 7:16649
Tripathi S, Sangwan RS, Mishra B, Jadaun JS, Sangwan NS (2019) Berry transcriptome: insights into a novel resource to understand development dependent secondary metabolism in Withania somnifera (Ashwagandha). Physiol Plant 168:148–173
Tripathi S, Srivastava Y, Sangwan RS, Sangwan NS (2020) In silico mining and functional analysis of AP2/ERF gene in Withania somnifera. Sci Rep 10:4877
Turck F, Zhou A, Somssich IE (2004) Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to its native promoter and the defense related gene PcPR1-1 in parsley. Plant Cell 16:2573–2585
Ülker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7:491–498
Vom Endt D, Kijne JW, Memelink J (2002) Transcription factors controlling plant secondary metabolism: what regulates the regulators? Phytochemistry 61:107–114
Wang JW, Wu JY (2010) Tanshinone biosynthesis in Salvia miltiorrhiza and production in plant tissue cultures. Appl Microbiol Biot 88:437–49
Wasternack C, Hause B (2002) Jasmonates and octadecanoids: signals in plant stress responses and development. Prog Nucleic Acid Res Mol Biol 72:165–221
Xu YH, Wang JW, Wang S, Wang JY, Chen XY (2004) Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-δ-cadinene synthase-A. Plant Physiol 135:507–515
Yamazaki Y, Sudo H, Yamazaki M, Aimi N, Saito K (2003) Camptothecin biosynthesis genes in hairy roots of Ophiorrhiza pumila: cloning, characterization and di verential expression in tissues and by stress compounds. Plant Cell Physiol 44:395–403
Zhang CQ, Xu Y, Lu Y, Yu HX, Gu MH, Liu QQ (2011) The WRKY transcription factor OsWRKY78 regulates stem elongation and seed development in rice. Planta 234:541–554
Acknowledgements
RSS and NSS thank New Millenium Indian Technology Leadership Initiative (NMITLI) for NMITLI Project grant on Ashwagandha. NSS also thanks Network project BSC 0203 for the grant for research and Dr V Gupta for providing vectors. JSJ acknowledges financial support of the University Grant Commission for research fellowship.
Funding
This work was supported by a Grant-in-Aid for research on Ashwagandha from New Millenium Indian Technology Leadership Initiative (NMITLI) for NMITLI Project.
Author information
Authors and Affiliations
Contributions
Experiments were designed, directed and conceived by RSS and NSS, and performed by JSJ, AKK and LKN. Manuscript writing and data analysis was done by RSS, NSS, SM and JSJ. All the authors approve the manuscript and declare no competing financial interacts.
Corresponding authors
Ethics declarations
Conflict of interest
No conflicts of interest declared.
Additional information
Communicated by Neal Stewart.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jadaun, J.S., Kushwaha, A.K., Sangwan, N.S. et al. WRKY1-mediated regulation of tryptophan decarboxylase in tryptamine generation for withanamide production in Withania somnifera (Ashwagandha). Plant Cell Rep 39, 1443–1465 (2020). https://doi.org/10.1007/s00299-020-02574-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-020-02574-4