Abstract
Key message
Combining phenotype and gene expression analysis of the CRISPR/Cas9-induced SlAN2 mutants, we revealed that SlAN2 specifically regulated anthocyanin accumulation in vegetative tissues in purple tomato cultivar ‘Indigo Rose.’
Abstract
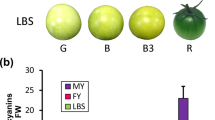
Anthocyanins play an important role in plant development and also exhibit human health benefits. The tomato genome contains four highly homologous anthocyanin-related R2R3-MYB transcription factors: SlAN2, SlANT1, SlANT1-like, and SlAN2-like/Aft. SlAN2-like/Aft regulates anthocyanin accumulation in the fruit; however, the genetic function of the other three factors remains unclear. To better understand the function of R2R3-MYB transcription factors, we conducted targeted mutagenesis of SlAN2 in the purple tomato cultivar ‘Indigo Rose’ using clustered regularly interspersed short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9). The SlAN2 mutants had a fruit color and anthocyanin content similar to cv. ‘Indigo Rose,’ while the anthocyanin content and the relative expression levels of several anthocyanin-related genes in vegetative tissues were significantly lower in the SlAN2 mutant relative to cv. Indigo Rose. Furthermore, we found that anthocyanin biosynthesis is controlled by different regulators between tomato hypocotyls and cotyledons. In addition, SlAN2 mutants were shorter, with smaller and lighter fruits than cv. ‘Indigo Rose.’ Our findings further our understanding of anthocyanin production in tomato and other plant species.





Similar content being viewed by others
References
Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE (2014) A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26:962–980
Boches P, Myers J (2007) The anthocyanin fruit tomato gene (Aft) is associated with a DNA polymorphism in a MYB transcription factor. In: HortScience. Amer Soc Horticultural Science 113 S WEST ST, STE 200, Alexandria, VA 22314–2851 USA, pp 856–856
Borevitz JO, Xia Y, Blount JW, Dixon RA, Lamb CJ (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12:2383–2393
Cao X, Qiu Z, Wang X, Van TG, Liu X, Wang J, Wang X, Gao J, Guo Y, Du Y (2017) A putative R3 MYB repressor is the candidate gene underlying atroviolacium, a locus for anthocyanin pigmentation in tomato fruit. J Exp Bot 68:5745–5758
Carrasco D, De Lorenzis G, Maghradze D, Revilla E, Bellido A, Failla O, Arroyogarcia R (2015) Allelic variation in the VvMYBA1 and VvMYBA2 domestication genes in natural grapevine populations (Vitis vinifera subsp. sylvestris). Plant Syst Evol 301:1613–1624
Chagne D, Carlisle CM, Blond C, Volz RK, Whitworth CJ, Oraguzie NC, Crowhurst RN, Allan AC, Espley RV, Hellens RP, Gardiner SE (2007) Mapping a candidate gene (MdMYB10) for red flesh and foliage colour in apple. BMC Genomics 8:212
Colanero S, Perata P, Gonzali S (2018) The atroviolacea gene encodes an R3-MYB protein repressing anthocyanin synthesis in tomato plants. Front Plant Sci 9:830
Colanero S, Tagliani A, Perata P, Gonzali S (2019) Alternative splicing in the anthocyanin fruit gene encoding an R2R3 MYB transcription factor affects anthocyanin biosynthesis in tomato fruits. Plant Commun 1:100006
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15:573–581
Elsm AA, Young JC, Rabalski I (2006) Anthocyanin composition in black, blue, pink, purple, and red cereal grains. J Agric Food Chem 54:4696–4704
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53:814–827
Gonzali S, Mazzucato A, Perata P (2009) Purple as a tomato: towards high anthocyanin tomatoes. Trends Plant Sci 14:237–241
Gould KS (2004) Nature's Swiss army knife: the diverse protective roles of anthocyanins in leaves. Biomed Res Int 2004:314–320
Hu D, Sun C, Ma Q, You C, Cheng L, Hao Y (2016) MdMYB1 regulates anthocyanin and malate accumulation by directly facilitating their transport into vacuoles in apples. Plant Physiol 170:1315–1330
Iwashina T (2015) Contribution to flower colors of flavonoids including anthocyanins: a review. Nat prod commun 10:529–544
Jaakola L (2013) New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci 18:477–483
Jeong S, Gotoyamamoto N, Hashizume K, Kobayashi S, Esaka M (2006) Expression of VvmybA1 gene and anthocyanin accumulation in various grape organs. Am J Enol Vitic 57:507–510
Jian W, Cao H, Yuan S, Liu Y, Lu J, Lu W, Li N, Wang J, Zou J, Tang N (2019) SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic Res 6:1–15
Kiferle C, Fantini E, Bassolino L, Povero G, Spelt C, Buti S, Giuliano G, Quattrocchio F, Koes R, Perata P (2015) Tomato R2R3-MYB proteins SlANT1 and SlAN2: same protein activity. Diff Roles PloS one 10:e0136365
Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, Mcwilliam H, Valentin F, Wallace IM, Wilm A, Lopez R (2007) Clustal W and clustal X version 2.0. Bioinformatics 23:2947–2948
Li Z, Peng R, Tian Y, Han H, Xu J, Yao Q (2016) Genome-wide identification and analysis of the MYB transcription factor superfamily in Solanum lycopersicum. Plant Cell Physiol 57:1657–1677
Liakopoulos G, Nikolopoulos D, Klouvatou A, Vekkos K-A, Manetas Y, Karabourniotis G (2006) The photoprotective role of epidermal anthocyanins and surface pubescence in young leaves of grapevine (Vitis vinifera). Ann Bot 98:257–265
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2—ΔΔCT method. Methods 25:402–408
Ma D, Constabel CP (2019) MYB repressors as regulators of phenylpropanoid metabolism in plants. Trends Plant Sci 24:275–289
Mathews H (2003) Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell Online 15:1689–1703
Mazza G (2007) Anthocyanins and heart health. Annali-Istituto Superiore Di Sanita 43:369
Meng X, Yang D, Li X, Zhao S, Sui N, Meng Q (2015) Physiological changes in fruit ripening caused by overexpression of tomato SlAN2, an R2R3-MYB factor. Plant Physiol Biochem 89:24–30
Nakabayashi R, Yonekura-Sakakibara K, Urano K, Suzuki M, Yamada Y, Nishizawa T, Matsuda F, Kojima M, Sakakibara H, Shinozaki K, Michael AJ, Tohge T, Yamazaki M, Saito K (2014) Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J 77:367–379
Nicholas K (1997) GeneDoc: analysis and visualization of genetic variation. Embnew News 4:28–30
Park SH, Morris JL, Park JE, Hirschi KD, Smith RH (2003) Efficient and genotype-independent Agrobacterium–mediated tomato transformation. J Plant Physiol 160:1253–1257
Paz-Ares J, Ghosal D, Wienand U, Peterson P, Saedler H (1987) The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. The EMBO J 6:3553–3558
Pireyre M, Burow M (2015) Regulation of MYB and bHLH transcription factors: a glance at the protein level. Mol Plant 8:378–388
Povero G, Gonzali S, Bassolino L, Mazzucato A, Perata P (2011) Transcriptional analysis in high-anthocyanin tomatoes reveals synergistic effect of Aft and atv genes. J Plant Physiol 168:270–279
Qiu Z, Wang H, Li D, Yu B, Hui Q, Yan S, Huang Z, Cui X, Cao B (2019) Identification of candidate HY5-dependent and -independent regulators of anthocyanin biosynthesis in tomato. Plant Cell Physiol 60:643–656
Qiu Z, Wang X, Gao J, Guo Y, Huang Z, Du Y (2016) The tomato hoffman's anthocyaninless gene encodes a bHLH transcription factor involved in anthocyanin biosynthesis that is developmentally regulated and induced by low temperatures. PloS one 11:e0151067
Rick C, Cisneros P, Chetelat R, DeVerna J (1994) Abg—a gene on chromosome 10 for purple fruit derived from S. lycopersicoides. Rep Tomato Genet Coop 44:29–30
Schreiber G, Reuveni M, Evenor D, Oren-Shamir M, Ovadia R, Sapir-Mir M, Bootbool-Man A, Nahon S, Shlomo H, Chen L, Levin I (2012) ANTHOCYANIN1 from Solanum chilense is more efficient in accumulating anthocyanin metabolites than its Solanum lycopersicum counterpart in association with the ANTHOCYANIN FRUIT phenotype of tomato. TAG Theoretical and applied genetics Theoretische und angewandte Genetik 124:295–307
Steyn W, Wand S, Holcroft D, Jacobs G (2002) Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytol 155:349–361
Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4:447–456
Sun C, Deng L, Du M, Zhao J, Chen Q, Huang T, Jiang H, Li C-B, Li C (2019) A transcriptional network promotes anthocyanin biosynthesis in tomato flesh. Mol Plant
Torres CA, Davies NM, Yanez JA, Andrews PK (2005) Disposition of selected flavonoids in fruit tissues of various tomato (Lycopersicon esculentum Mill.) genotypes. J Agric Food Chem 53:9536–9543
Vimolmangkang S, Han Y, Wei G, Korban SS (2013) An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol 13:176
Xu W, Dubos C, Lepiniec L (2015) Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci 20:176–185
Yan S, Chen N, Huang Z, Li D, Zhi J, Yu B, Liu X, Cao B, Qiu Z (2019) Anthocyanin fruit encodes an R2R3-MYB transcription factor, SlAN2-like, activating the transcription of SlMYBATV to fine-tune anthocyanin content in tomato fruit. New Phytol 225:2048–2063
Yousuf B, Gul K, Wani AA, Singh P (2016) Health benefits of anthocyanins and their encapsulation for potential use in food systems: a review. Crit Rev Food Sci Nutr 56:2223–2230
Yu B, Yan S, Zhou H, Dong R, Lei J, Chen C, Cao B (2018) Overexpression of CsCaM3 improves high temperature tolerance in cucumber. Front Plant Sci 9:797
Zhang Y, Butelli E, De Stefano R, Schoonbeek H-j, Magusin A, Pagliarani C, Wellner N, Hill L, Orzaez D, Granell A (2013) Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Curr Biol 23:1094–1100
Zhang Y, de Stefano R, Robine M, Butelli E, Bulling K, Hill L, Rejzek M, Martin C, Schoonbeek H-j (2015) Different ROS-scavenging properties of flavonoids determine their abilities to extend shelf life of tomato. Plant Physiol 86:S11
Zhou H, Linwang K, Wang F, Espley RV, Ren F, Zhao J, Ogutu C, He H, Jiang Q, Allan AC (2019) Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol 221:1919–1934
Zhu H, Zhang T-J, Zheng J, Huang X-D, Yu Z-C, Peng C-L, Chow WS (2018) Anthocyanins function as a light attenuator to compensate for insufficient photoprotection mediated by nonphotochemical quenching in young leaves of Acmena acuminatissima in winter. Photosynthetica 56:445–454
Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40:22–34
Acknowledgements
This study was supported by the National Natural Science Foundation of China (31801863) and the Natural Science Foundation of Guangdong Province (2018A030310211).
Author information
Authors and Affiliations
Contributions
ZQ and BC conceived and designed the experiments; JZ and XL performed majority of experiments and data analyses. DL, YH, BY, SY, and BC performed experiments and data analyses; ZQ and XL wrote the manuscript; SY and BC provided input and revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare that they have no competing interests.
Additional information
Communicated by Kan Wang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhi, J., Liu, X., Li, D. et al. CRISPR/Cas9-mediated SlAN2 mutants reveal various regulatory models of anthocyanin biosynthesis in tomato plant. Plant Cell Rep 39, 799–809 (2020). https://doi.org/10.1007/s00299-020-02531-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-020-02531-1