Abstract
Key message
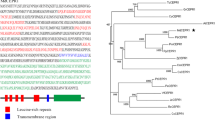
MdCRY2 was isolated from apple fruit skin, and its function was analyzed in MdCRY2 transgenic Arabidopsis. The interaction between MdCRY2 and AtCOP1 was found by yeast two-hybrid and BiFC assays.
Abstract
Cryptochromes are blue/ultraviolet-A (UV-A) light receptors involved in regulating various aspects of plant growth and development. Investigations of the structure and functions of cryptochromes in plants have largely focused on Arabidopsis (Arabidopsis thaliana), tomato (Solanum lycopersicum), pea (Pisum sativum), and rice (Oryza sativa). However, no data on the function of CRY2 are available in woody plants. In this study, we isolated a cryptochrome gene, MdCRY2, from apple (Malus domestica). The deduced amino acid sequences of MdCRY2 contain the conserved N-terminal photolyase-related domain and the flavin adenine dinucleotide (FAD) binding domain, as well as the C-terminal DQXVP-acidic-STAES (DAS) domain. Relationship analysis indicates that MdCRY2 shows the highest similarity to the strawberry FvCRY protein. The expression of MdCRY2 is induced by blue/UV-A light, which represents a 48-h circadian rhythm. To investigate the function of MdCRY2, we overexpressed the MdCRY2 gene in a cry2 mutant and wild type (WT) Arabidopsis, assessed the phenotypes of the resulting transgenic plants, and found that MdCRY2 functions to regulate hypocotyl elongation, root growth, flower initiation, and anthocyanin accumulation. Furthermore, we examined the interaction between MdCRY2 and AtCOP1 using a yeast two-hybrid assay and a bimolecular fluorescence complementation assay. These data provide functional evidence for a role of blue/UV-A light-induced MdCRY2 in controlling photomorphogenesis in apple.






Similar content being viewed by others
References
Ahmad M, Cashmore AR (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366:162–166
Allan AC, Hellens RP, Laing WA (2008) MYB transcription factors that colour our fruit. Trends Plant Sci 13:99–102
Briggs WR, Christie JM (2002) Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci 7(5):204–210
Brudler R, Hitomi K, Daiyasu H, Toh H, Kucho K, Ishiura M, Kanehisa M, Roberts VA, Todo T, Tainer JA, Getzoff ED (2003) Identification of a new cryptochrome class. Structure, function, and evolution. Mol Cell 11:59–67
Canamero RC, Bakrim N, Bouly JP, Garay A, Dudkin EE, Habricot Y, Ahmad M (2006) Cryptochrome photoreceptors cry1 and cry2 antagonistically regulate primary root elongation in Arabidopsis thaliana. Planta 224:995–1003
Cashmore AR (2003) Cryptochromes: enabling plants and animals to determine circadian time. Cell 114:537–543
Chatterjee M, Sharma P, Khurana JP (2006) Cryptochrome1 from Brassica napus is up-regulated by blue light and controls hypocotyl/stem growth and anthocyanin accumulation. Plant Physiol 141:61–74
Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, O’Hara A, Kelly SM, Hothorn M, Smith BO, Hitomi K, Jenkins KI, Getzoff ED (2012) Plant UVR8 photo-receptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 335:1492–1496
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Correll MJ, Kiss JZ (2005) The roles of phytochromes in elongation and gravitropism of roots. Plant Cell Physiol 46(2):317–323
Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49:414–427
Franklin KA, Larner VS, Whitelam GC (2005) The signal transducing photoreceptors of plants. Int J Dev Biol 49:653–664
Guo H, Yang H, Mockler TC, Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279:1360–1363
Gyula P, Schafer E, Nagy F (2003) Light perception and signaling in higher plants. Curr Opin Plant Biol 6:446–452
Hong SH, Kim HJ, Ryu JS, Choi H, Jeong S, Shin J, Choi G, Nam HG (2008) CRY1 inhibits COP1-mediated degradation of BIT1, a MYB transcription factor, to activate blue light-dependent gene expression in Arabidopsis. Plant J 55:361–371
Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27:1277–1288
Kleine T, Lockhart P, Batschauer A (2003) An Arabidopsis protein closely related to Synechocystis cryptochrome is targeted to organelles. Plant J 35(1):93–103
Li YY, Mao K, Zhao C, Zhao XY, Zhang HL, Shu HR, Hao YJ (2012) MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol 160:1011–1022
Lin C (2000) Plant blue-light receptors. Trends Plant Sci 5(8):337–342
Lin C (2002) Blue light receptors and signal transduction. Plant Cell 14:207–225
Lin C, Shalitin D (2003) Cryptochrome structure and signal transduction. Annu Rev Plant Biol 54:469–496
Lin C, Ahmad M, Gordon D, Cashmore A (1995) Expression of an Arabidopsis cryptochrome gene in transgenic tobacco results in hypersensitivity to blue, UV-A, and green light. Proc Natl Acad Sci USA 92:8423–8427
Lin C, AhmadM Chan J, Cashmore AR (1996) CRY2, a second member of the Arabidopsis cryptochrome gene family. Plant Physiol 110:1047
Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR (1998) Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA 95:2686–2690
Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20:292–306
Liu H, Liu B, Zhao C, Pepper M, Lin C (2011) The action mechanisms of plant cryptochromes. Trends Plant Sci 16(12):684–691
Mandoli DF, Ford GA, Waldron LJ, Nemson JA, Briggs WR (1990) Some spectral properties of several soil types: implications for photomorphogenesis. Plant Cell Environ 13:287–294
Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405:462–466
Partch CL, Sancar A (2005) Cryptochromes and circadian photoreception in animals. Methods Enzymol 393:726–745
Platten JD, Foo E, Elliott RC, Hecht V, Reid JB, Weller JL (2005) Cryptochrome 1 contributes to blue-light sensing in pea. Plant Physiol 139:1472–1482
Quattrocchio F, Wing J, Woude K, Souer E, Vetten N, Mol J, Koes R (1999) Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11(8):1433–1444
Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schäfer E, Nagy F, Jenkins GI, Ulm R (2011) Perception of UV-B by the Arabidopsis UVR8 protein. Science 332:103–106
Sakamoto K, Briggs WR (2002) Cellular and subcellular localization of phototropin1. Plant Cell 14(8):1723–1735
Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423:995–999
Toth R, Kevei E, Hall A, Millar AJ, Nagy F, Kozma-Bognar L (2001) Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol 127:1607–1616
Wang H, Ma LG, Li JM, Zhao HY, Deng XW (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294:154–158
Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA (2003) The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol 20(9):1377–1419
Wu G, Spalding EP (2007) Separate functions for nuclear and cytoplasmic cryptochrome 1 during photomorphogenesis of Arabidopsis seedlings. Proc Natl Acad Sci USA 104:18813–18818
Wu D, Hu Q, Yan Z, Chen W, Yan C, Huang X, Zhang J, Yang P, Deng H, Wang J, Deng XW, Shi Y (2012) Structural basis of ultraviolet-B perception by UVR8. Nature 484:214–219
Xu P, Xiang Y, Zhu H, Xu H, Zhang Z, Zhang C, Zhang L, Ma Z (2009) Wheat cryptochromes: subcellular localization and involvement in photomorphogenesis and osmotic stress responses. Plant Physiol 149:760–774
Yang HQ, Tang RH, Cashmore AR (2001) The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13:2573–2587
Yang J, Lin R, Sullivan J, Hoecker U, Liu B, Xu L, Deng XW, Wang H (2005) Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17:804–821
Yu X, Shalitin D, Liu X, Maymon M, Klejnot J, Yang H, Lopez J, Zhao X, Bendehakkalu KT, Lin C (2007) Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc Natl Acad Sci USA 104:7289–7294
Yu X, Sayegh R, Maymon M, Warpeha K, Klejnot J, Yang H, Huang J, Lee J, Kaufman L, Lin C (2009) Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation. Plant Cell 21:118–130
Yu X, Liu H, Klejnot J, Lin C (2010) The cryptochrome blue-light receptors. The Arabidopsis book 8:e0135. doi:10.1199/tab.0135
Zhang Q, Li H, Li R, Hu R, Fan C, Chen F, Wang Z, Liu X, Fu Y, Lin C (2008) Association of the circadian rhythmic expression of GmCRY1a with a latitudinal cline in photoperiodic flowering of soybean. Proc Natl Acad Sci USA 105:21028–21033
Acknowledgments
We thank Prof. Hongquan Yang of Shanghai Jiao Tong University, China, for providing Arabidopsis cry2 mutant. This work was supported by National Basic Research Program of China (2011CB100600), National Natural Science Foundation of China (31272142), and Program for Changjiang Scholars and Innovative Research Team in University (IRT1155).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Chong.
Electronic supplementary material
Below is the link to the electronic supplementary material.
299_2013_1387_MOESM1_ESM.tif
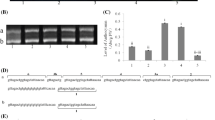
Figure S1. Expression of AtPAP1, AtBIT1, AtCO, AtHY5, AtCHS, and AtDFR in WT and MdCRY2 overexpressing Arabidopsis. AtACTIN was used as the external control (TIFF 112 kb)
Rights and permissions
About this article
Cite this article
Li, YY., Mao, K., Zhao, C. et al. Molecular cloning and functional analysis of a blue light receptor gene MdCRY2 from apple (Malus domestica). Plant Cell Rep 32, 555–566 (2013). https://doi.org/10.1007/s00299-013-1387-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-013-1387-4