Abstract
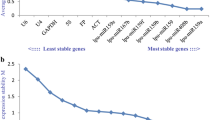
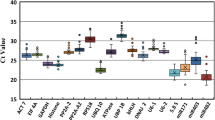
miRNAs have recently been reported to modulate somatic embryogenesis (SE), a key pathway of plant regeneration in vitro. For expression level detection and subsequent function dissection of miRNAs in certain biological processes, qRT-PCR is one of the most effective and sensitive techniques, for which suitable reference gene selection is a prerequisite. In this study, three miRNAs and eight non-coding RNAs (ncRNA) were selected as reference candidates, and their expression stability was inspected in developing citrus SE tissues cultured at 20, 25, and 30 °C. Stability of the eight non-miRNA ncRNAs was further validated in five adult tissues without temperature treatment. The best single reference gene for SE tissues was snoR14 or snoRD25, while for the adult tissues the best one was U4; although they were not as stable as the optimal multiple references snoR14 + U6 for SE tissues and snoR14 + U5 for adult tissues. For expression normalization of less abundant miRNAs in SE tissues, miR3954 was assessed as a viable reference. Single reference gene snoR14 outperformed multiple references for the overall SE and adult tissues. As one of the pioneer systematic studies on reference gene identification for plant miRNA normalization, this study benefits future exploration on miRNA function in citrus and provides valuable information for similar studies in other higher plants.
Key message Three miRNAs and eight non-coding RNAs were tested as reference candidates on developing citrus SE tissues. Best single references snoR14 or snoRD25 and optimal multiple references snoR14 + U6, snoR14 + U5 were identified.





Similar content being viewed by others
Abbreviations
- miRNA:
-
microRNA
- ncRNA:
-
Non-coding RNA
- qRT-PCR:
-
Quantitative reverse transcription-polymerase chain reaction
- SE:
-
Somatic embryogenesis
- NEC:
-
Non-embryogenic callus
- EC:
-
Embryogenic callus
- E1/2/3/4:
-
Embryogenic callus induced for somatic embryo for 1/2/3/4 weeks
- GE:
-
Globular embryo
- HE:
-
Heart embryo
- CE:
-
Cotyledonary embryo
- Ct:
-
Cycle threshold
- Csi:
-
Citrus sinensis
- TPM:
-
Times per million
References
Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Borges A, Tsai SM, Caldas DGG (2012) Validation of reference genes for RT-qPCR normalization in common bean during biotic and abiotic stresses. Plant Cell Rep 31:827–838
Brattelid T, Aarnes EK, Helgeland E, Guvag S, Eichele H, Jonassen AK (2011) Normalization strategy is critical for the outcome of miRNA expression analyses in the rat heart. Physiol Genomics 43:604–610
Buhtz A, Pieritz J, Springer F, Kehr J (2010) Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol 10:64
Carra A, Mica E, Gambino G, Pindo M, Moser C, Pè ME, Schubert A (2009) Cloning and characterization of small non-coding RNAs from grape. Plant J 59:750–763
Carvalho K, de Campos MKF, Pereira LFP, Vieira LGE (2010) Reference gene selection for real-time quantitative polymerase chain reaction normalization in “Swingle” citrumelo under drought stress. Anal Biochem 402:197–199
Chandna R, Augustine R, Bisht NC (2012) Evaluation of candidate reference genes for gene expression normalization in Brassica juncea using real time quantitative RT-PCR. PLoS One 7:e36918
Chang KH, Mestdagh P, Vandesompele J, Kerin MJ, Miller N (2010) MiRNA expression profiling to identify and validate reference genes for relative quantification in colorectal cancer. BMC Cancer 10:173
Chen XM (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 35:21–44
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33(20):e179
Davoren PA, McNeill RE, Lowery AJ, Kerin MJ, Miller N (2008) Identification of suitable endogenous control genes for microRNA gene expression analysis in human breast cancer. BMC Mol Biol 9:76
de Vega-Bartol J, Santos RR, Simões M, Miguel C (2011) Evaluation of reference genes for quantitative PCR analysis during somatic embryogenesis in conifers. BMC Proc 5(Suppl 7):O44
Deng XX (1987) Studies on the isolation, regeneration and fusion of protoplasts in Citrus. Ph.D Dissertation of Huazhong Agricultural University
Ding YF, Chen Z, Zhu C (2011) Microarray-based analysis of cadmium-responsive microRNAs in rice (Oryza sativa). J Exp Bot 62:3563–3573
Galiveti CR, Rozhdestvensky TS, Brosius J, Lehrach H, Konthur Z (2010) Application of housekeeping npcRNAs for quantitative expression analysis of human transcriptome by real-time PCR. RNA 16:450–461
Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, Bateman A (2009) Rfam: updates to the RNA families database. Nucleic Acids Res 37:D136–D140
Gimenez MJ, Piston F, Atienza SG (2011) Identification of suitable reference genes for normalization of qPCR data in comparative transcriptomics analyses in the Triticeae. Planta 233:163–173
Gmitter FG Jr, Chen CX, Machado MA, de Souza AA, Ollitrault P, Froehlicher Y, Shimizu T (2012) Citrus genomics. Tree Genet Genomes 8:611–626
Goncalves S, Cairney J, Maroco J, Oliveira MM, Miguel C (2005) Evaluation of control transcripts in real-time RT-PCR expression analysis during maritime pine embryogenesis. Planta 222:556–563
Guo WW, Li DL, Duan YX (2007) Citrus transgenics: current status and prospects. Transgenic Plant J 1:202–209
Gutierrez L, Mauriat M, Pelloux J, Bellini C, Van Wuytswinkel O (2008a) Towards a systematic validation of references in real-time RT-PCR. Plant Cell 20:1734–1735
Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Van Wuytswinkel O (2008b) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol 6:609–618
Jang JS, Vernadette AS, Feddersen RM, Rakhshan F, Schultz DA, Zschunke MA, Lingle WL, Kolbert CP, Jen J (2011) Quantitative miRNA expression analysis using fluidigm microfluidics dynamic arrays. BMC Genomics 12:144
Kulcheski FR, Marcelino-Guimaraes FC, Nepomuceno AL, Abdelnoor RV, Margis R (2010) The use of microRNAs as reference genes for quantitative polymerase chain reaction in soybean. Anal Biochem 406:185–192
Kulcheski FR, de Oliveira LF, Molina LG, Almerão MP, Rodrigues FA, Marcolino J, Barbosa JF, Stolf-Moreira R, Nepomuceno AL, Marcelino-Guimarães FC, Abdelnoor RV, Nascimento LC, Carazzolle MF, Pereira GAG, Margis R (2011) Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genomics 12:307
Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, Calin GA, Ivan M (2006) A microRNA signature of hypoxia. Mol Cell Biol 27:1859–1867
Kumar P, Johnston BH, Kazakov SA (2011) miR-ID: a novel, circularization-based platform for detection of microRNAs. RNA 17:365–380
Lalitha S (2000) Primer Premier 5. Biotech Softw Internet Rep 1:270–272
Lim QE, Zhou L, Ho YK, Wan G, Too HP (2011) snoU6 and 5S RNAs are not reliable miRNA reference genes in neuronal differentiation. Neuroscience 199:32–43
Lin YL, Lai ZX (2010) Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Sci 178:359–365
Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, Alder H, Bullrich F, Negrini M, Croce CM (2004) An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA 101:9740–9744
Liu Y, Liu Q, Tao NG, Deng XX (2006) Efficient isolation of RNA from fruit peel and pulp of ripening navel orange (Citrus sinensis Osbeck). J Huazhong Agric Univ 25:300–304
Liu Q, Xu J, Liu Y, Zhao X, Deng X, Guo L, Gu J (2007) A novel bud mutation that confers abnormal patterns of lycopene accumulation in sweet orange fruit (Citrus sinensis L. Osbeck). J Exp Bot 58:4161–4171
Mafra V, Kubo KS, Alves-Ferreira M, Ribeiro-Alves M, Stuart RM, Boava LP, Rodrigues CM, Machado MA (2012) Reference genes for accurate transcript normalization in Citrus genotypes under different experimental conditions. PLoS One 7(2):e31263
Maroufi A, Van Bockstaele E, De Loose M (2010) Validation of reference genes for gene expression analysis in chicory (Cichorium intybus) using quantitative real-time PCR. BMC Mol Biol 11:15
Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, Vandesompele J (2009) A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol 10:R64
Meyer SU, Pfaffl MW, Ulbrich SE (2010) Normalization strategies for microRNA profiling experiments: a ‘normal’ way to a hidden layer of complexity? Biotechnol Lett 32:1777–1788
Murashige T, Tucker DPH (1969) Growth factor requirement of citrus tissue cultures. In: Chapman HD (ed) Proceedings of the International Citrus Symposium, vol 3. Riverside, California, pp 1155–1161
Nakano M, Shimada T, Endo T, Fujii H, Nesumi H, Kita M, Ebina M, Shimizu T, Omura M (2012) Characterization of genomic sequence showing strong association with polyembryony among diverse Citrus species and cultivars, and its synteny with Vitis and Populus. Plant Sci 183:131–142
Ollitrault P, Guo WW, Grosser JW (2007) Somatic hybridization. In: Khan I (ed) Citrus genetics, breeding and biotechnology. CABI, pp 235–260
Peltier HJ, Latham GJ (2008) Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA 14:844–852
Peng T, Lv Q, Zhang J, Li JZ, Du YX, Zhao QZ (2011) Differential expression of the microRNAs in superior and inferior spikelets in rice (Oryza sativa). J Exp Bot 62:4943–4954
Pérez RM (2000) Cryostorage of Citrus embryogenic cultures. In: Jain SM, Gupta PK, Newton RJ (eds) Somatic embryogenesis in woody plants, vol 6. Kluwer, Dordrecht, pp 687–705
Schaefer A, Jung M, Miller K, Lein M, Kristiansen G, Erbersdobler A, Jung K (2010) Suitable reference genes for relative quantification of miRNA expression in prostate cancer. Exp Mol Med 42:749–758
Selim M, Legay S, Berkelmann-Löhnertz B, Langen G, Kogel KH, Evers D (2012) Identification of suitable reference genes for real-time RT-PCR normalization in the grapevine-downy mildew pathosystem. Plant Cell Rep 31:205–216
Shen JQ, Xie KB, Xiong LZ (2010) Global expression profiling of rice microRNAs by one-tube stem-loop reverse transcription quantitative PCR revealed important roles of microRNAs in abiotic stress responses. Mol Genet Genomics 284:477–488
Shen YM, Li Y, Ye F, Wang FF, Wan XY, Lu WG, Xie X (2011) Identification of miR-23a as a novel microRNA normalizer for relative quantification in human uterine cervical tissues. Exp Mol Med 43:358–366
Shi R, Chiang VL (2005) Facile means for quantifying microRNA expression by real-time PCR. Biotechniques 39:519–525
Song CN, Wang C, Zhang CQ, Korir NK, Yu HP, Ma3 ZQ, Fang JG (2010) Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata). BMC Genomics 11:431
Tong ZG, Gao ZH, Wang F, Zhou J, Zhang Z (2009) Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol Biol 10:71
Udvardi MK, Czechowski T, Scheible WR (2008) Eleven golden rules of quantitative RT-PCR. Plant Cell 20:1736–1737
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:research 0034.1–0034.11
Varkonyi-Gasic E, Wu R, Wood M, Walton E, Hellens R (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3:12
Wang TZ, Chen L, Zhao MG, Tian QY, Zhang WH (2011) Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high throughput sequencing. BMC Genomics 12:367
Wang C, Han J, Liu CH, Kibet KN, Kayesh E, Shangguan LF, Li XY, Fang JG (2012) Identification of microRNAs from Amur grape (vitis amurensis Rupr.) by deep sequencing and analysis of microRNA variations with bioinformatics. BMC Genomics 13:122
Wessels JM, Edwards AK, Zettler C, Tayade C (2011) Selection and validation of reference genes for miRNA expression studies during porcine pregnancy. PLoS One 6(12):e28940
Wu XM, Liu MY, Ge XX, Xu Q, Guo WW (2011) Stage and tissue-specific modulation of ten conserved miRNAs and their targets during somatic embryogenesis of Valencia sweet orange. Planta 233:495–505
Xu Q, Liu Y, Zhu A, Wu X, Ye J, Yu K, Guo W, Deng XX (2010) Discovery and comparative profiling of microRNAs in a sweet orange red-flesh mutant and its wild type. BMC Genomics 11:246
Yan JW, Yuan FR, Long GY, Qin L, Deng ZN (2012) Selection of reference genes for quantitative real-time RT-PCR analysis in citrus. Mol Biol Rep 39:1831–1838
Zhong HY, Chen JW, Li CQ, Chen L, Wu JY, Chen JY, Lu WJ, Li JG (2011) Selection of reliable reference genes for expression studies by reverse transcription quantitative real-time PCR in litchi under different experimental conditions. Plant Cell Rep 30:641–653
Acknowledgments
This research was financially supported by the Ministry of Science & Technology of China (nos. 2011CB100606, 2011AA100205), the National NSF of China and the Doctoral Research Fund of Ministry of Education of China (no. 20100146110008). The authors thank Dr. Bo Zheng (Huazhong Agric. Univ.) for reading the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Rose.
S.-J. Kou and X.-M Wu contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kou, SJ., Wu, XM., Liu, Z. et al. Selection and validation of suitable reference genes for miRNA expression normalization by quantitative RT-PCR in citrus somatic embryogenic and adult tissues. Plant Cell Rep 31, 2151–2163 (2012). https://doi.org/10.1007/s00299-012-1325-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-012-1325-x