Abstract
Establishment of the shoot apical meristem (SAM) in Arabidopsis embryos requires the KNOXI transcription factor SHOOT MERISTEMLESS. In Norway spruce (Picea abies), four KNOXI family members (HBK1, HBK2, HBK3 and HBK4) have been identified, but a corresponding role in SAM development has not been demonstrated. As a first step to differentiate between the functions of the four Norway spruce HBK genes, we have here analyzed their expression profiles during the process of somatic embryo development. This was made both under normal embryo development and under conditions of reduced SAM formation by treatment with the polar auxin transport inhibitor NPA. Concomitantly with the formation of an embryonic SAM, the HBK2 and HBK4 genes displayed a significant up-regulation that was delayed by NPA treatment. In contrast, HBK1 and HBK3 were up-regulated prior to SAM formation, and their temporal expression was not affected by NPA. Ectopic expression of the four HBK genes in transgenic Arabidopsis plants further supported similar functions of HBK2 and HBK4, distinct from those of HBK1 and HBK3. Together, the results suggest that HBK2 and HBK4 exert similar functions related to the SAM differentiation and somatic embryo development in Norway spruce, while HBK1 and HBK3 have more general functions during embryo development.
Similar content being viewed by others
Introduction
The shoot apical meristem (SAM) is established early during plant embryogenesis. It includes a group of self-renewing cells, which eventually give rise to all above-ground parts of a plant. The regulation of establishment and maintenance of the SAM has been thoroughly studied in the angiosperm Arabidopsis thaliana. However, angiosperms and gymnosperms separated approximately 300 million years ago (Smith et al. 2010), and our knowledge about the corresponding processes in gymnosperms is limited.
The establishment of the embryonal SAM in Arabidopsis is dependent on the expression of the homeodomain containing transcription factor SHOOT MERISTEMLESS (STM; Barton and Poethig 1993; Long et al. 1996). STM is one of four class I KNOTTED1-like homeobox (KNOXI) genes in Arabidopsis, the other members being KNOTTED-like from Arabidopsis thaliana 1/BREVIPEDICELLUS (KNAT1/BP), KNAT2 and KNAT6. KNOXI genes are expressed in the SAM and down-regulated before leaf initiation. In addition, they are expressed in the margins of compound leaves, where they presumably prevent cell differentiation (Hay and Tsiantis 2010). STM is the best studied gene within the KNOXI gene family. Since the other members act redundantly with STM in the development of both vegetative and reproductive meristematic tissues, it has been difficult to assign specific functions to each gene (Hay and Tsiantis 2010).
STM is the first KNOXI gene to be expressed during embryogenesis. It marks the entire SAM, and its expression is essential for establishment of the embryonal SAM (Barton and Poethig 1993; Long et al. 1996). KNAT6 is also expressed in the embryonal SAM, but not until bilateral symmetry has been established (Belles-Boix et al. 2006). It marks the SAM boundaries and acts redundantly with STM to maintain meristem identity and organ separation (Belles-Boix et al. 2006). BP is expressed in the shoot apex at the seedling stage (Dockx et al. 1995). There is no disruption of the SAM in bp mutants (Venglat et al. 2002), although BP can restore the SAM function when ectopically expressed in stm mutants (Scofield et al. 2008). Mutants of knat2 have no obvious phenotype, probably because of redundancy with KNAT6 (Byrne et al. 2002). However, ectopically expressed KNAT2 induces a conversion of ovules to carpels, suggesting a function related to carpel development (Pautot et al. 2001), and also STM has been shown to be important for carpel development (Scofield et al. 2007). The delineation of embryonal shoot and root meristems in Arabidopsis is correlated with auxin maxima, established by polar auxin transport (PAT; Möller and Weijers 2009). We have previously shown that PAT also is of major importance for the correct patterning of the embryonal shoot and root in Norway spruce (Larsson et al. 2008). To further characterize the molecular regulation of SAM establishment in conifers, it is important to know more about the expression of KNOXI genes during SAM formation.
Four KNOXI genes, HBK1, HBK2, HBK3 and PaKN4 (from now on denoted HBK4 for simplicity), have been identified in Norway spruce (Guillet-Claude et al. 2004; Hjortswang et al. 2002; Sundås-Larsson et al. 1998). HBK1, HBK2 and HBK3 are expressed in embryogenic cultures, stems, roots, and female and male cone buds, but not in needles (Hjortswang et al. 2002). In addition, HBK1 shows a tissue-specific expression in vegetative meristems (Sundås-Larsson et al. 1998). Overexpression of HBK3 leads to enlarged SAMs in somatic embryos and an accelerated differentiation of early embryos from proembryonic masses (PEMs), while down-regulation of HBK3 precludes embryo differentiation (Belmonte et al. 2007). Furthermore, it has been shown that HBK2 is expressed only in embryogenic cell lines that are competent to form fully mature cotyledonary embryos, while HBK1 and HBK3 are expressed also in developmentally arrested lines (Hjortswang et al. 2002). However, there are so far no reports on the expression pattern of HBK4.
Phylogenetic analyses have shown that the four KNOXI genes in spruce form a monophyletic group and likely have diversified after the split between angiosperms and gymnosperms (Guillet-Claude et al. 2004). The authors suggested that gene losses have occurred in conifers after new paralogs were gained and further proposed that HBK1 and HBK3 would exert similar and redundant functions, while HBK2 and HBK4 should be more diverse.
Here, we have analyzed the expression of all four HBK genes during Norway spruce embryo development and correlated their expression to PAT inhibition and the establishment of a functional SAM. The expression profiles, together with expression studies in transgenic Arabidopsis plants, indicate that HBK1 and HBK3 act in similar pathways, while HBK2 and HBK4 have several features in common, but are distinct from HBK1 and HBK3. Furthermore, the temporal expression of HBK2 and HBK4 during early embryo development suggests functions connected to SAM establishment and somatic embryo formation.
Materials and methods
Plant materials
The embryogenic cell line 28:05 of Norway spruce (Picea abies L. Karst) was used throughout this study. It was stored in liquid nitrogen and thawed approximately 6 months before the start of the experiments. After thawing, the cell cultures were treated as described previously (von Arnold and Clapham 2008). Briefly, proembryogenic masses (PEMs) were maintained in liquid proliferation medium containing the plant growth regulators (PGRs), 2,4-dichlorophenoxyacetic acid (2,4-D) and N 6-benzyladenine (BA) at 10.0 and 4.4 µM, respectively. The cultures were transferred to fresh medium weekly. To stimulate differentiation of early somatic embryos from PEMs, the cultures were transferred to pre-maturation medium lacking PGRs for 1 week. For development of late somatic embryos and maturation, the cultures were plated on solidified maturation medium containing 30 µM of abscisic acid (ABA).
To study the effect of PAT on the expression of the HBK genes, embryogenic cultures were treated with 20 µM 1-N-naphthylphthalamic acid (NPA; Sigma-Aldrich, Schnelldorf, Germany) during both the pre-maturation and maturation phases, as described by Larsson et al. (2008). Samples for gene expression studies were collected from eight consecutive stages from both control and NPA-treated embryos (Fig. 1). The developmental stages of NPA-treated embryos were largely defined according to the sizes of phenotypically normal control embryos. Whole mount of tissue was sampled from proliferating PEMs (stage 1) and early embryos (stage 2). From stage 3 and onwards, the embryos were sampled individually and sorted according to their developmental stage to increase the specificity in subsequent expression analyses. Samples were stored at −80°C until use.
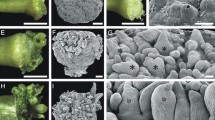
Eight consecutive stages of control (a–d, h–k) and NPA-treated (e–g, l–o) somatic embryos of Norway spruce. The developmental stages of NPA-treated embryos were largely defined according to the sizes of phenotypically normal control embryos, and not according to age; however, the average exposure time to ABA for reaching a certain stage is presented. a Stage 1, proliferating proembryonic masses in the presence of the plant growth regulators (PGRs) auxin and cytokinin. NPA was added to the cultures simultaneously with the withdrawal of PGRs, and stage 1 cultures were hence not treated with NPA. b, e Stage 2, early embryos 1 week after withdrawal of PGRs. c, f Stage 3, beginning of late embryo development after 1–2 weeks of exposure to ABA. Note the cone shape of the embryo treated with NPA (f) compared to the more cylindrical shape of the control embryo (c). d, g Stage 4, late embryos after 2–3 weeks of exposure to ABA. Note that the suspensor is still present in the NPA-treated embryo (g), while it has been degraded in the control embryo (d). h, l Stage 5, early maturing embryos after 3–4 weeks of exposure to ABA. i, m Stage 6, maturing embryos after 4–5 weeks (i) or 5–6 weeks (m) of exposure to ABA. j, n Stage 7, almost fully matured control embryo after 5–6 weeks of exposure to ABA (j), almost fully matured embryo with doughnut-shaped apical part after 6–7 weeks of exposure to ABA and NPA (n). k, o Stage 8, fully matured cotyledonary embryo after 6–7 weeks of exposure to ABA (k), and fully matured embryo lacking separated cotyledons after 7–8 weeks of exposure to ABA and NPA (o). co Cotyledon, cp cotyledon primordia, dsc doughnut-shaped cotyledon, em embryonal mass, fc fused cotyledons, s suspensor, sp shoot apical meristem primordium, SAM shoot apical meristem, tc tube cells. Scale bars 100 µm (a–j, l–n) and 250 µm (k, o)
Cell line 88:1, in which proliferation of PEMs continue on pre-maturation medium (Hjortswang et al. 2002), was used to analyze the expression of the HBK genes in an arrested embryogenic cell line. Samples were taken after 1 week of proliferation in the presence of PGRs, after 1 week in pre-maturation medium, and after 1 week on maturation medium. Samples from cell line 28:05 were used as control.
RNA isolation and cDNA synthesis
For quantitative real-time PCR (qRT-PCR), total RNA was extracted according to a modified protocol from Azevedo et al. (2003). Briefly, samples snap frozen in liquid nitrogen were disrupted in TissueLyZer II (Retsch, Haan, Germany). Extraction buffer was added to each sample, which was again frozen in liquid nitrogen, thawed at 42°C, disrupted and finally incubated at 42°C for 90 min, followed by subsequent RNA isolation. For real-time PCR (RT-PCR), total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the protocol provided by the manufacturer.
An aliquot (10 µg) of each sample was treated with DNase using the DNA-free protocol by Ambion (Ambion inc, Austin, TX, USA), and 1 µg of the DNA-free RNA was used to synthesise cDNA using the qScript™ cDNA Synthesis Kit (Quanta BioSciences, Inc. Gaithersburg, MD, USA) according to the protocol provided by the manufacturer.
Quantitative real-time PCR
qRT-PCR was performed using the DyNAmo™ Flash SYBR® Green qPCR Kit (Finnzymes, Espoo, Finland) in a BIO-RAD iQ™5 Multicolor Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Primers used to quantify expression levels are presented in Online Resource Suppl. Tab. 1a. Expression data were normalized against the expression of the reference genes CELL DIVISION CONTROL2 (CDC2), ELONGATION FACTOR-1 ALFA (EF1-α) and PHOSPHOGLUCOMUTASE, previously selected based on their stability (Vestman et al. 2011) as determined using the geNorm software (Vandesompele et al. 2002). PCR cycling conditions were as advised by the manufacturer in the DyNAmo™ Flash SYBR® Green qPCR Kit (Finnzymes, Espoo, Finland) with annealing and extension at 60°C for 30 s. The reactions were run for 40 cycles followed by the generation of a melting curve to ensure product uniformity. All samples were added in triplicate to each plate, and each gene was measured in three independent biological replicates. The analyses were performed using the iQ5 software (Bio-Rad Laboratories, Hercules, CA, USA).
Statistical analyses of the results from the qRT-PCR were performed using the SAS (2008) software general linear model (GLM) procedure. The effects of treatment and sample on each gene expression level were analyzed using GLM with treatment and sample as factors. The different genes were analyzed separately.
Vector construction for overexpression in Arabidopsis
Full-length cDNAs of HBK2, HBK3 and HBK4 were amplified from early somatic embryos of Norway spruce, and full-length cDNA of HBK1 was amplified from a pBluescript clone kindly provided by Dr. Annika Sundås-Larsson (Uppsala University, Uppsala, Sweden) using primers designed from published sequences (Accession numbers AF063248, AF483277, AF483278, AY680389 and AY680400). Primers are presented in Online Resource Suppl. Tab. 1b. Amplified products were subcloned into the pJET1.2/blunt cloning vector using the CloneJET™ PCR Cloning Kit (Fermentas Helsingborg, Sweden). All cDNA clones were subsequently re-amplified using primers with attB-adapters and inserted into the 35S promoter containing Gateway vector pGWB2 (Nakagawa et al. 2007) using the Gateway® technology according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA). Primers are presented in Online Resource Suppl. Tab. 1c. The resulting vectors were introduced by freeze–thawing into Agrobacterium tumefaciens strain C58:C1. Arabidopsis plants, ecotype Columbia (Col), were transformed using the floral-dip method and transgenic seedlings were selected on kanamycin-containing plates. Resistant seedlings were planted in soil and phenotypically analyzed.
Results and discussion
We have previously shown that formation of a functional SAM during embryo development in Norway spruce is dependent on PAT (Larsson et al. 2008). Briefly, treatment with the well-established PAT inhibitor NPA delayed the development of somatic embryos, and mature somatic embryos showed abnormal morphology with fused or aborted cotyledons and no histologically visible SAM. These embryos could neither germinate nor develop an epicotyl, indicating lack of a functional root meristem and SAM.
A model system for studying genes correlated with SAM formation in conifer embryos
To analyze the expression of the four HBK genes during embryo development in Norway spruce, and whether the expression was different in embryos lacking a functional SAM, both control somatic embryos and somatic embryos treated with NPA were separated into eight consecutive developmental stages (Fig. 1). This classification increases the resolution during late embryogeny and maturation compared to our previous work (Larsson et al. 2008).
Proliferating PEMs were considered as stage 1 (Fig. 1a). Early embryos that had started to differentiate from the larger PEM structures represented stage 2 (Fig. 1b). At stage 3 (Fig. 1c), the embryos had developed further and consisted of an embryonal mass with a smooth surface and a long suspensor made up of highly vacuolated cells. The embryonal mass and the suspensor were separated by tube cells, which are specific for gymnosperms. Stage 4 (Fig. 1d) was represented by late embryos with a dense embryonal mass, which was continuously growing while most of the suspensor cells had been degraded by programmed cell death, a feature that is essential for the formation of viable somatic embryos (Bozhkov et al. 2005; Smertenko et al. 2003). Stage 5 (Fig. 1h) was characterized by maturing embryos, which had clearly visible cotyledon primordia surrounding a convex SAM primordium. At this stage, all suspensor cells had been degraded. During stages 6–8 (Fig. 1i–k), the embryos continued to increase in size, the cotyledons, which could be seen as small protuberances at stage 6 (Fig. 1i), expanded further at stage 7 (Fig. 1j) and finally fully spread out at stage 8 (Fig. 1k).
The development of NPA-treated embryos was divided into eight consecutive stages mainly defined according to the size of control embryos. There were no distinct differences between control embryos and NPA-treated embryos until the cotyledon primordia were clearly visible (cf. Fig. 1b–g). However, compared to the cylindrical control embryos (Fig. 1b, c), NPA-treated embryos often carried more suspensor cells, resulting in cone-shaped early embryos (Fig. 1e, f). In general, the NPA-treated embryos developed more slowly and the suspensor did not degrade at the same rate as for control embryos. As the embryos matured, it became evident that the cotyledons were fused, giving the apex a doughnut appearance when observed from above (Fig. 1n). At stage 8, when the cotyledons of control embryos burst, the doughnut thinned out at the edges and there was a deep cavity at the center (Fig. 1o). We have previously shown by histological analysis that the SAM is missing in such embryos (Larsson et al. 2008).
Expression of HBK2 and HBK4 coincides with the SAM formation
The relative expression level of HBK1, HBK2, HBK3 and HBK4 was analyzed by quantitative real-time PCR in both control embryos and in embryos that had been treated with NPA throughout their development. Since we have previously shown that NPA treatment of PEMs does not affect SAM formation (Larsson et al. 2008), stage 1 cultures remained untreated. Thus, the expression of each gene at each stage and treatment could be related to the expression at stage 1.
HBK1 was expressed in proliferating PEMs and the expression was significantly up-regulated from stage 3 and then down-regulated in mature embryos (Fig. 2a). The relative expression of HBK3 (Fig. 2c) was similar to that of HBK1, although HBK3 became significantly up-regulated first at stage 4 and then remained at a high expression level. Treatment with NPA did not significantly alter the expression of either gene. These results show that the accumulation of both HBK1 and HBK3 mRNA precedes the differentiation of the SAM. However, since NPA-treated embryos lack a visible SAM (Larsson et al. 2008), the absence of an NPA-response on HBK1 and HBK3 expression suggests that the genes are not specifically related to SAM establishment, and thus have a more general role in embryo development.
Quantitative real-time PCR analysis of HBK1 (a), HBK2 (b), HBK3 (c) and HBK4 (d) mRNA levels during eight consecutive stages of control (dark bars) and NPA-treated (light bars) somatic embryos of Norway spruce. Embryos from stage 3 and later were sampled individually. Expression values are relative to the expression of each gene at stage 1, normalized against three reference genes. The expression levels are mean values of three biological replicates, each analyzed in triplicate and presented in a logarithmic scale. Error bars indicate SEM of biological replicates. Asterisks indicate a difference between stage 1 and later stages of control cultures, significant at *p < 0.05 or **p < 0.01. Circles indicate a significant difference (p < 0.01) between control and NPA-treated embryos at the stages indicated
In contrast, HBK2 was expressed in proliferating PEMs, but as early embryos started to differentiate at stage 2, the expression decreased and reached its lowest level at stage 3 and 4 (Fig. 2b). At stage 5, when the cotyledons started to develop and the SAM primordium was bulging out from the center of the cotyledon crown, the HBK2 expression increased drastically and the mRNA level was 20 times higher compared to that at stage 3 and 4. From stage 6, the relative expression of HBK2 was two to three times that compared to stage 1, and almost 40 times higher as compared to that at stage 3 and 4. Compared to control embryos, treatment with NPA led to a significantly higher HBK2 expression at stage 2, but a significantly lower expression at stage 5 and 6. Taken together, the expression of HBK2 started to increase between stages 4 and 5 in control embryos, coinciding with the appearance of the SAM primordium, while it was not until stage 6 when the fused cotyledon ring became visible that HBK2 expression increased in NPA-treated embryos. Interestingly, NPA treatment did not alter the expression of HBK2 when all stages were analyzed as a group, indicating that NPA treatment delayed the up-regulation of HBK2. The delay in HBK2 expression could either be a direct effect of the blocked PAT, or it could mirror an indirect effect, where up-stream regulators of HBK2 are directly affected by the blocked PAT. It has been shown in tomato (Solanum lycopersicum) that there is a narrow window during leaf development when the leaf is responsive to KNOXI activity (Shani et al. 2009). Assuming a similar spatial and temporal window during embryo development in Norway spruce, our results suggest that HBK2 must be up-regulated at stage 5 for an appropriate differentiation of SAM.
The relative expression level of HBK4 (Fig. 2d) was low in proliferating PEMs, but increased as embryo morphogenesis began. At stage 5, when the SAM was visible at the center of the small cotyledon protuberances, HBK4 was significantly up-regulated compared to stage 1, and the expression level then remained high throughout the maturation process. There was a statistically significant difference between the HBK4 mRNA accumulation in NPA-treated embryos and in control embryos. The expression of HBK4 was initially down-regulated between stages 1 and 2 in NPA-treated embryos. Furthermore, the up-regulation of HBK4 observed in control embryos at stages 5 and 6, when SAM and cotyledon differentiation began, was partly inhibited in NPA-treated embryos. It was not until the embryos were mature that the relative expression level became comparable in NPA-treated and control embryos. These results suggest that also HBK4 should be up-regulated at a certain time point for the SAM to be established.
The qRT-PCR results show that the highly similar genes, HBK1 and HBK3, are expressed in a similar way during embryo development, and that their temporal expression levels are not affected in embryos lacking a functional SAM. This suggests that HBK1 and HBK3 have more general roles during embryo development, rather than specific functions during embryonal SAM establishment. In contrast, both HBK2 and HBK4 become up-regulated during the establishment of the embryonal SAM. This temporal up-regulation is comparable to that of STM in Arabidopsis (Long et al. 1996), indicating that HBK2 and HBK4 have roles in SAM establishment. In addition, the increase in the relative expression of HBK2 and HBK4 is delayed in NPA-treated embryos lacking a SAM, which support that appropriate timing of the expression of HBK2 and HBK4 is important for SAM establishment.
HBK2 and HBK4 are specifically expressed in cell lines competent to form fully mature cotyledonary embryos
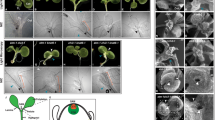
It has been shown that HBK1 and HBK3 are expressed both in embryogenic cell lines that form fully mature cotyledonary embryos, and in blocked cell lines that never pass the PEM-to-embryo transition stage (Hjortswang et al. 2002). This is in contrast to HBK2, which is only expressed in embryogenic cell lines that form fully mature embryos. To further investigate the similarities and differences between the four HBK genes, the temporal expression of HBK4 was compared to that of HBK1, HBK2 and HBK3 in a control line (28:05) and in a blocked cell line (88:1). HBK1 and HBK3 were expressed in both cell lines, while HBK2 and HBK4 were expressed only in cell line 28:05 (Fig. 3). These results support a similar regulation of HBK1 and HBK3, and of HBK2 and HBK4. The results also reveal an intriguing, albeit tentative, link between the expression of HBK2 and HBK4 and proper embryo maturation, thus providing potential markers for somatic embryo competence.
RT-PCR analysis of HBK1, HBK2, HBK3 and HBK4 expression in embryogenic cultures that were either competent to form fully mature cotyledonary embryos (28:05) or blocked at the PEM-to-embryo transition stage (88:1). Cell lines were sampled after 1 week of proliferation in the presence of PGRs (1), after 1 week in pre-maturation medium (2) and after 1 week on maturation medium (3). For cell line 28:05, these stages represent stages 1–3 in Fig. 1. The expression of PHOSPHOGLUCOMUTASE was used as a reference and H2O was used as a negative control in the RT-PCR analysis
Constitutive expression of HBK1 and HBK3 in Arabidopsis induces morphologies characteristic for ectopic KNOXI expression
Constitutive expression of KNAT1/BP and STM, as well as of different KNOXI genes from other plant species, induces ectopic meristems on leaves (Hay and Tsiantis 2010). Hence, it was of interest to analyze if any of the HBK genes also could mimic this feature. Thus, each of the four HBK genes were expressed from the 35S promoter in transgenic Arabidopsis plants. It should be noted that expressing conifer genes in an angiosperm such as Arabidopsis does not necessarily provide information on the precise gene function in conifers, although phenotyping Arabidopsis plants constitutively expressing either of the four HBK genes may indicate if the genes have similar or distinct functions in spruce.
The expression of all four HBK genes induced variable degrees of serrated and lobed rosette and cauline leaves (Table 1, Online Resource Fig. 1b–e), typical for KNOXI overexpressors (Hay and Tsiantis 2010 with refs). In general, HBK1 and HBK3 induced more severely serrated leaves, while most of the HBK2 and HBK4 transformants only showed mild serration. In addition, the plants expressing HBK1 and HBK3 had abnormal flowers (Table 1, Online Resource Fig. 1g–o), similar to BP, KNAT2 and STM overexpressors (Scofield et al. 2008). The flowers of HBK2 and HBK4 expressing plants were morphologically similar to wild-type flowers (Online Resource Fig. 1h, j), although the abscission of the outer floral organs after flowering was extremely delayed in the most severe HBK2 and HBK4 lines (Online Resource Fig. 1m, o). Surprisingly, none of the spruce genes induced ectopic SAMs on the Arabidopsis leaf surfaces. The results show that HBK genes have some features in common with angiosperm KNOXI genes when constitutively expressed in Arabidopsis. However, since the spruce genes did not induce SAM differentiation, we assume that HBK genes are too diverse compared to angiosperm KNOXI genes to be able to stimulate such induction. In addition, the delayed abscission of the HBK2 and HBK4 expressing flowers is a characteristic, not previously presented for KNOXI overexpressors. This suggests that HBK2 and HBK4 can function in pathways other than those previously described for KNOXI proteins.
Taken together, our data show that HBK1 and HBK3 have similar expression profiles during somatic embryo differentiation and development in Norway spruce. Although HBK1 and HBK3 can function in pathways similar to angiosperm KNOXI genes when expressed in Arabidopsis, their temporal expression patterns during somatic embryogenesis in Norway spruce suggest that HBK1 and HBK3 have a more general function during embryo development than to specify the SAM. In contrast, HBK2 and HBK4 are specifically expressed in embryogenic cell lines competent to develop into fully mature cotyledonary embryos, and their temporal expression profiles are correlated with the formation of the embryonal SAM. This suggests that HBK2 and HBK4 are essential for somatic embryogenesis and the formation of a functional SAM in Norway spruce.
References
Azevedo H, Lino-Neto T, Tavares RM (2003) An improved method for high-quality RNA isolation from needles of adult maritime pine trees. Plant Mol Biol Rep 21:333–338
Barton MK, Poethig RS (1993) Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development 119:823–831
Belles-Boix E, Hamant O, Witiak SM, Morin H, Traas J, Pautot V (2006) KNAT6: an Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell 18:1900–1907
Belmonte M, Tahir M, Schroeder D, Stasolla C (2007) Overexpression of HBK3, a class I KNOX homeobox gene, improves the development of Norway spruce (Picea abies) somatic embryos. J Exp Bot 58:2851–2861
Bozhkov PV, Filonova LH, Suarez MF (2005) Programmed cell death in plant embryogenesis. Curr Top Dev Biol 67:135–179
Byrne ME, Simorowski J, Martienssen RA (2002) ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129:1957–1965
Dockx J, Quaedvlieg N, Keultjes G, Kock P, Weisbeek P, Smeekens S (1995) The homeobox gene ATK1 of Arabidopsis thaliana is expressed in the shoot apex of the seedling and in flowers and inflorescence stems of mature plants. Plant Mol Biol 28:723–737
Guillet-Claude C, Isabel N, Pelgas B, Bousquet J (2004) The evolutionary implications of knox-I gene duplication in conifers: correlated evidence from phylogeny, gene mapping, and analysis of functional divergence. Mol Biol Evol 21:2232–2245
Hay A, Tsiantis M (2010) KNOX genes: versatile regulators of plant development and diversity. Development 137:3153–3165
Hjortswang HI, Sundås Larsson A, Bharathan G, Bozhkov PV, von Arnold S, Vahala T (2002) KNOTTED1-like homeobox genes of a gymnosperm, Norway spruce, expressed during somatic embryogenesis. Plant Physiol Biochem 40:837–843
Larsson E, Sitbon F, Ljung K, von Arnold S (2008) Inhibited polar auxin transport results in aberrant embryo development in Norway spruce. New Phytol 177:356–366
Long JA, Maon EI, Medford JI, Barton MK (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379:66–69
Möller B, Weijers D (2009) Auxin control of embryo patterning. Cold Spring Harbor Perspect Biol 1:a001545
Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104:34–41
Pautot V, Dockx J, Hamant O, Kronenberger J, Grandjean O, Jublot D, Traas J (2001) KNAT2: evidence for a link between Knotted-like genes and carpel development. Plant Cell 13:1719–1734
Scofield S, Dewitte W, Murray JAH (2007) The KNOX gene SHOOT MERISTEMLESS is required for the development of reproductive meristematic tissues in Arabidopsis. Plant J 50:767–781
Scofield S, Dewitte W, Murray JAH (2008) A model for Arabidopsis class-1 KNOX gene function. Plant Signal Behav 3:257–259
Shani E, Burko Y, Ben-Yaakov L, Berger Y, Amsellem Z, Goldshmidt A, Sharon E, Ori N (2009) Stage-specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell 21:3078–3092
Smertenko AP, Bozhkov PV, Filonova LH, von Arnold S, Hussey PJ (2003) Re-organisation of the cytoskeleton during developmental programmed cell death in Picea abies embryos. Plant J 33:813–824
Smith SA, Beaulieu JM, Donoghue MJ (2010) An uncorrelated relaxed-clock analysis suggests an earlier origin for flowering plants. Proc Natl Acad Sci USA 107:5897–5902
Sundås-Larsson A, Svenson M, Liao H, Engström P (1998) A homeobox gene with potential developmental control function in the meristem of the conifer Picea abies. Proc Natl Acad Sci USA 95:15118–15122
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7):research 0034.1–0034.11
Venglat SP, Dumonceaux T, Rozwadowski K, Parnell L, Babic V, Keller W, Martienssen R, Selvaraj G, Datla R (2002) The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc Natl Acad Sci USA 99:4730–4735
Vestman D, Larsson E, Uddenberg D, Cairney J, Clapham D, Sundberg E, von Arnold S (2011) Important processes during differentiation and early development of somatic embryos of Norway spruce as revealed by changes in global gene expression. Tree Genet Genomes 7:347–362
von Arnold S, Clapham D (2008) Spruce embryogenesis. In: Suárez MF, Bozhkov PV (eds) Plant embryogenesis methods in molecular biology. Humana, Totowa, pp 31–47
Acknowledgments
We thank Ulf Olsson for statistical analyses and Gunilla Swärdh at the Uppsala Arabidopsis transformation platform for technical assistance. This work was supported by the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Jordan.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Larsson, E., Sitbon, F. & von Arnold, S. Differential regulation of Knotted1-like genes during establishment of the shoot apical meristem in Norway spruce (Picea abies). Plant Cell Rep 31, 1053–1060 (2012). https://doi.org/10.1007/s00299-011-1224-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-011-1224-6