Abstract
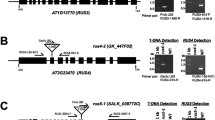
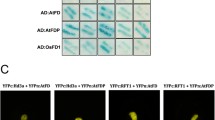
DWF4 encodes a rate-limiting mono-oxygenase that mediates 22α-hydroxylation reactions in the BR biosynthetic pathway and it is the target gene in the BR feedback loop. Knockout of DWF4 results in a dwarfed phenotype and other severe defects in Arabidopsis. Here we report on the isolation of the ZmDWF4 gene in maize. Sequence analysis revealed that the open reading frame of ZmDWF4 was 1,518 bp, which encodes a protein composed of 505 amino acid residues with a calculated molecular mass of 57.6 kD and a predicated isoelectric point (pI) of 9.54. Phylogenetic analysis indicated that ZmDWF4 was very close to the Arabidopsis DWF4. In young maize seedlings, the expression of ZmDWF4 in shoots was much higher than that in roots. The highest expression of ZmDWF4 was observed in husk leaves and the lowest in silks during flowering stage. The expression of ZmDWF4 in maize was significantly down regulated by exogenous brassinolide. A heterogeneous complementary experiment demonstrated that the defects of three Arabidopsis DWF4 mutants could be rescued by constitutive expression of ZmDWF4, with leaf expandability, inflorescence stem heights and fertile capabilities all restored to normal levels. Increases in seed and branch number as well as the height of florescence stem were observed in the over-expressed transformants. These findings suggest that ZmDWF4 may be an ortholog gene of Arabidopsis DWF4 and responsible for BR biosynthesis in maize.





Similar content being viewed by others
References
Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301:653–657
Altmann T (1998) Recent advances in brassinosteroid molecular genetics. Curr Opin Plant Biol 1:378–383
Asami T, Nakano T, Fujioka S (2005) Plant brassinosteroid hormones. Vitam Horm 72:479–504
Azpiroz R, Wu Y, LoCascio JC, Feldmann KA (1998) An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell 10:219–230
Castle J, Szekeres M, Jenkins G, Bishop GJ (2005) Unique and overlapping expression patterns of Arabidopsis CYP85 genes involved in brassinosteroid C-6 oxidation. Plant Mol Biol 57:129–140
Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA (1998) The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22alpha-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10:231–243
Choe S, Dilkes BP, Gregory BD, Ross AS, Yuan H, Noguchi T, Fujioka S, Takatsuto S, Tanaka A, Yoshida S, Tax FE, Feldmann KA (1999) The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol 119:897–907
Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA (2001) Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J 26:573–582
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Clouse SD, Sasse JM (1998) BRASSINOSTEROIDS: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49:427–451
Dean C, Elzen PV, Tamaki S, Dunsmuir P, Bedbrook J (1985) Differential expression of the eight genes of the petunia ribulose bisphosphate carboxylase small subunit multi-gene family. Embo J 4:3055–3061
Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19:1349
Felsenstein J (1989) PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 5:164–166
Fujioka S, Yokota T (2003) Biosynthesis and metabolism of brassinosteroids. Annu Rev Plant Biol 54:137–164
Fujita S, Ohnishi T, Watanabe B, Yokota T, Takatsuto S, Fujioka S, Yoshida S, Sakata K, Mizutani M (2006) Arabidopsis CYP90B1 catalyses the early C-22 hydroxylation of C27, C28 and C29 sterols. Plant J 45:765–774
Grove M, Spencer GF, Rohwedder WK (1979) Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature 281:216–217
He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307:1634–1638
Kim GT, Fujioka S, Kozuka T, Tax FE, Takatsuto S, Yoshida S, Tsukaya H (2005) CYP90C1 and CYP90D1 are involved in different steps in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J 41:710–721
Klahre U, Noguchi T, Fujioka S, Takatsuto S, Yokota T, Nomura T, Yoshida S, Chua NH (1998) The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell 10:1677–1690
Li L, Xu J, Xu ZH, Xue HW (2005) Brassinosteroids stimulate plant tropisms through modulation of polar auxin transport in Brassica and Arabidopsis. Plant Cell 17:2738–2753
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods Mol Biol 25:402–408
Mussig C (2005) Brassinosteroid-promoted growth. Plant Biol (Stuttg) 7:110–117
Nebert DW, Gonzalez FJ (1987) P450 genes: structure,evolution, and regulation. Annu Rev Biochem 56:945–993
Nemhauser JL, Chory J (2004) BRing it on: new insights into the mechanism of brassinosteroid action. J Exp Bot 55:265–270
Noguchi T, Fujioka S, Takatsuto S, Sakurai A, Yoshida S, Li J, Chory J (1999) Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-En-3-one to (24R)-24-methyl-5alpha-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol 120:833–840
Nomura T, Kushiro T, Yokota T, Kamiya Y, Bishop GJ, Yamaguchi S (2005) The last reaction producing brassinolide is catalyzed by cytochrome P-450s, CYP85A3 in tomato and CYP85A2 in Arabidopsis. J Biol Chem 280:17873–17879
Retief JD (2000) Phylogenetic analysis using PHYLIP. Methods Mol Biol 132:243–258
Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, Tanaka H, Kitano H, Matsuoka M (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol 24:105–109
Shi YH, Zhu SW, Mao XZ, Feng JX, Qin YM, Zhang L, Cheng J, Wei LP, Wang ZY, Zhu YX (2006) Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18:651–664
Shimada Y, Fujioka S, Miyauchi N, Kushiro M, Takatsuto S, Nomura T, Yokota T, Kamiya Y, Bishop GJ, Yoshida S (2001) Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol 126:770–779
Shimada Y, Goda H, Nakamura A, Takatsuto S, Fujioka S, Yoshida S (2003) Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol 131:287–297
Symons GM, Reid JB (2004) Brassinosteroids do not undergo long-distance transport in pea. Implications for the regulation of endogenous brassinosteroid levels. Plant Physiol 135:2196–2206
Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85:171–182
Takahashi N, Nakazawa M, Shibata K, Yokota T, Ishikawa A, Suzuki K, Kawashima M, Ichikawa T, Shimada H, Matsui M (2005) shk1-D, a dwarf Arabidopsis mutant caused by activation of the CYP72C1 gene, has altered brassinosteroid levels. Plant J 42:13–22
Tanaka K, Asami T, Yoshida S, Nakamura Y, Matsuo T, Okamoto S (2005) Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol 138:1117–1125
Tao Yz, Zheng J, Xu Zm, Zhang Xh, Zhang K, Wang GY (2004) Functional analysis of ZmDWF1, a maize homolog of the Arabidopsis brassinosteroids biosynthetic DWF1/DIM gene. Plant Sci 167:743–751
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Wang H, Torres QI, Ward JM, Murthy G, Zhang J, Walker JC, Neff MM (2005) BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms. Plant J 42:23–34
Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, Chory J (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2:505–513
Wang ZY, Wang Q, Chong K, Wang F, Wang L, Bai M, Jia C (2006) The brassinosteroid signal transduction pathway. Cell Res 16:427–434
Acknowledgments
This work was supported by the National High-tech Program of China (2006AA10A106).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Jordan.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, T., Zhang, J., Wang, M. et al. Expression and functional analysis of ZmDWF4, an ortholog of Arabidopsis DWF4 from maize (Zea mays L.). Plant Cell Rep 26, 2091–2099 (2007). https://doi.org/10.1007/s00299-007-0418-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-007-0418-4