Abstract
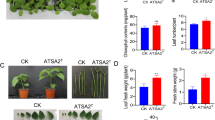
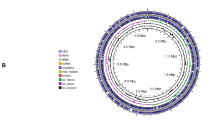
Plant growth-promoting (PGP) bacteria are an environmental-friendly alternative to chemical fertilizers for promoting plant development. We isolated and characterized a PGP endophyte, YSD J2, from the leaves of Cyperus esculentus L. var. sativus. Specific PGP characteristics of this strain, such as phosphate solubilization ability, potassium-dissolving ability, siderophore and indole-3-acetic acid (IAA) production, and salt tolerance, were determined in vitro. In addition, positive mutants were screened using the atmospheric and room-temperature plasma (ARTP) technology, with IAA level and organic phosphorus solubility as indices. Furthermore, the effect of the positive mutant on biomass production and antioxidant abilities of greengrocery seedling was evaluated and the genome was mined to explore the underlying mechanisms. The strain YSD J2 showed a good performance of PGP characteristics, such as the production of indole acetic acid and siderophores, solubilization ability of phosphate, and potassium-dissolving ability. It was recognized through 16S rRNA sequencing together with morphological and physiological tests and confirmed as Pantoea sp. The strain exposed to a mutation time of 125 s by ARTP had the highest IAA and organic phosphate (lecithin) concentrations of 10.34 mg/L and 16.52 mg/L, 42.06% and 34.15% higher than those of the initial strain. Inoculation of mutant strain YSD J2 significantly increased plant growth attributes and the activities of peroxidase and superoxide dismutase, respectively, but decreased the content of malondialdehyde significantly compared with the control. Furthermore, genome annotation and functional analysis were performed through whole-genome sequencing and PGP-related genes were identified. Our results indicated that the YSD J2 with PGP characteristics is a potential candidate for the development of biofertilizers.





Similar content being viewed by others
Abbreviations
- C. esculentus L.:
-
Cyperus esculentus L. var. sativus Baeck
- PGP:
-
Plant growth-promoting
- IAA:
-
Indole-3-acetic acid
- ARTP:
-
Atmospheric and room-temperature plasma
- MR:
-
Methyl red
- VP:
-
Voges–Proskauer
- SOD:
-
Superoxide dismutase
- POD:
-
Peroxidase
- MDA:
-
Malondialdehyde
- TCA:
-
Trichloroacetic acid
- SMRT:
-
Single-molecule real-time
- CDSs:
-
Coding sequences
- GI:
-
Gene Island
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- COG:
-
Clusters of Orthologous Group
- NR:
-
Non-redundant protein
- CAZy:
-
Carbohydrate-active enzymes
- PHI:
-
Pathogen–host interactions
- CARD:
-
Comprehensive antibiotic resistance database
- NRPS:
-
Non-ribosomal peptide synthase
- ANOVA:
-
Analysis of variance
References
Huygh W, Larridon I, Reynders M, Muasya AM, Govaerts R, Simpson DA, Goetghebeur P (2010) Nomenclature and typification of names of genera and subdivisions of genera in Cypereae (Cyperaceae): 1. Names of genera in the Cyperus clade. Taxon 59:1883–1890
Larridon I, Huygh W, Reynders M, Muasya AM, Govaerts R, Simpson DA, Goetghebeur P (2011) Nomenclature and typification of names of genera and subdivisions of genera in Cypereae (Cyperaceae): 2. Names of subdivisions of Cyperus. Taxon 60:868–884
Nyarko HD, Daniel NAT, Yaw A (2011) Assessment of microbiological safety of tiger nuts (Cyperus esculentus L.) in the Cape Coast Metropolis of Ghana. Arch Appl Sci Res 3(6):257–262
Bado S, Bazongo P, Son G, Kyaw MT, Bassolé IHN (2015) Physicochemical characteristics and composition of three morphotypes of cyperus esculentus tubers and tuber oils. J Anal Methods Chem 2015:1–8
Ntukidem VE, Ukwo PS, Udoh IE, Umoinyang EU (2019) Influence of different pretreatments on the nutritional and organoleptic properties of vegetable milk produced locally from tiger-nut (Cyperus esculentus) tubers. IOSR-JESTFT 13(6):55–61
Jing SQ, Wang SS, Li Q, Zheng L, Yue L, Fan SL, Tao GJ (2016) Dynamic high pressure microfluidization-assisted extraction and bioactivities of Cyperus esculentus (C. esculentus L.) leaves flavonoids. Food Chem 192:319–327
Jing SQ, Wang SS, Zhong RM, Zhang JY, Wu JZ, Tu YX, Pu Y, Yan LJ (2020) Neuroprotection of Cyperus esculentus L. orientin against cerebral ischemia/reperfusion induced brain injury. Neural Regen Res 15(03):548–556
Calvo P, Nelson L, Kloepper JW (2014) Agricultural uses of plant biostimulants. Plant Soil 383:3–41
Vinale F, Sivasithamparam K, Ghisalberti E, Marra R, Woo S, Lorito M (2007) Trichoderma-plant-pathogen interactions. Soil Biol Biochem 40:1–10
Santoyo G, Moreno-Hagelsieb G, Orozco-Mosqueda M, Glick B (2016) Plant growth-promoting bacterial endophytes. Microbiol Res 83:92–99
Strobel G, Stierle A, Stierle D, Hess WM (1993) Taxomyces andreanae, a proposed new taxon for a Bulbilliferous hyphomycete associated with Pacific yew (Taxus brevifolia). Mycotaxon (USA) 47(71):71–80
Kang SM, Radhakrishnan R, You YH, Joo GJ, Lee IJ, Lee KE, Kim JH (2014) Phosphate solubilizing Bacillus megaterium mj1212 regulates endogenous plant carbohydrates and amino acids contents to promote mustard plant growth. Indian J Microbiol 54:427–433
Ryu CM, Farag MA, Hu CH, Reddy MS, Wei HX, Pare PW, Kloepper JW (2003) Bacterial volatiles promote growth in arabidopsis. Proc Natl Acad Sci USA 100:4927–4932
Wilson MK, Abergel RJ, Raymond KN, Arceneaux JEL, Byers BR (2006) Siderophores of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. Biochem Biophys Res Commun 348:320–325
Morales-Cedeño LR, del Orozco-Mosqueda MC, Loeza-Lara PD, Parra-Cota FI, de los Santos-Villalobos S, Santoyo G (2021) Plant growth-promoting bacterial endophytes as biocontrol agents of pre- and post-harvest diseases: fundamentals, methods of application and future perspectives. Microbiol Res. https://doi.org/10.1016/j.micres.2020.126612
Yang L, Zhou N, Tian Y (2019) Characterization and application of dextranase produced by Chaetomium globosum mutant through combined application of atmospheric and room temperature plasma and ethyl methyl sulfone. Process Biochem 85:116–124
Gao XL, Liu E, Yin YY, Yang LX, Huang QR, Chen S, Ho CT (2020) Enhancing activities of salt-tolerant proteases secreted by Aspergillus oryzae using atmospheric and room-temperature plasma mutagenesis. J Agric Food Chem 68:2757–2764
Shu L, Si X, Yang X, Ma W, Sun J, Zhang J, Xue X, Wang D, Gao Q (2020) Enhancement of acid protease activity of Aspergillus oryzae using atmospheric and room temperature plasma. Front Microbiol 11:1418
Ottenheim C, Nawrath M, Wu JC (2018) Microbial mutagenesis by atmospheric and room-temperature plasma (ARTP): the latest development. Bioresour Bioprocess 5(1):1–12
Chen L, Shi H, Heng JY, Wang DX, Bian K (2019) Antimicrobial, plant growth-promoting and genomic properties of the peanut endophyte Bacillus velezensis LDO2. Microbiol Res 218:41–48
Wang W, Wu Z, He Y, Huang Y, Li X, Ye BC (2018) Plant growth promotion and alleviation of salinity stress in Capsicum annuum L. by Bacillus isolated from saline soil in Xinjiang. Ecotoxicol Environ Saf 164:520–529
Nanjundan J, Ramasamy R, Uthandi S, Ponnusamy M (2019) Antimicrobial activity and spectroscopic characterization of surfactin class of lipopeptides from Bacillus amyloliquefaciens SR1. Microb Pathogenesis 128:374–380
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Mukhtar S, Shahid I, Mehnaz S, Malik KA (2017) Assessment of two carrier materials for phosphate solubilizing biofertilizers and their effect on growth of wheat (Triticum aestivum L.). Microbiol Res 205:107–117
Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56
Yaghoubi Khanghahi M, Pirdashti H, Rahimian H, Nematzadeh G, Ghajar Sepanlou M (2018) Potassium solubilising bacteria (KSB) isolated from rice paddy soil: from isolation, identification to K use efficiency. Symbiosis 76:13–23. https://doi.org/10.1007/s13199-017-0533-0
Ramesh A, Sharma SK, Sharma MP, Yadav N, Joshi OP (2014) Inoculation of zinc solubilizing Bacillus aryabhattai strains for improved growth, mobilization and biofortification of zinc in soybean and wheat cultivated in Vertisols of central India. Appl Soil Ecol 73:87–96
Ahmad F, Ahmad I, Khan MS (2008) Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol Res 163(2):173–181
Hmaeid N, Wali M, Metoui-Ben Mahmoud O, Pueyo JJ, Ghnaya T, Abdelly C (2019) Efficient rhizobacteria promote growth and alleviate NaCl-induced stress in the plant species Sulla carnosa. Appl Soil Ecol 133:104–113
Zhuang Y, Jiang GL, Zhu MJ (2020) Atmospheric and room temperature plasma mutagenesis and astaxanthin production from sugarcane bagasse hydrolysate by Phaffia rhodozyma mutant Y1. Process Biochem 91:330–338
Fan X, Wu H, Li G, Yuan H, Zhang H, Li Y (2017) Improvement of uridine production of Bacillus subtilis by atmospheric and room temperature plasma mutagenesis and high-throughput screening. PLoS ONE 12(5):e0176545
Shi L, Thinn NT, Ge B, Zhao W, Liu B, Cui H, Zhang K (2018) Antifungal and plant growth-promoting activities of Streptomyces roseoflavus strain NKZ-259. Biol Control 125:57–64
Ullah I, Waqas M, Khan MA (2017) Exogenous ascorbic acid mitigates flood stress damages of Vigna angularis. Appl Biol Chem 60:603–614
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Faino L, Seidl MF, Datema E, van den Berg GC, Janssen A, Wittenberg AH, Thomma BP (2015) Single-molecule real-time sequencing combined with optical mapping yields completely finished fungal genome. MBio 6:e00936-00915
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Wang J (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1:18
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD (2012) Spades: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5):455–477
Besemer J, Borodovsky M (2005) GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res 33:W451–W454
Delcher AL, Bratke KA, Powers EC, Salzberg SL (2007) Identifying bacterial genes and endosymbiont DNA with GLIMMER. Bioinformatics 23(6):673–679
Chan PP, Lowe TM (2019) tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol 1962:1–14
Bertelli C, Laird MR, Williams KP (2017) IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res 45:W30
Fouts DE (2006) Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res 34(20):5839–5851
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28(1):27–30
Jensen LJ, Julien P, Kuhn M, Mering CV, Muller J, Doerks T, Bork P (2008) eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res 36(suppl 1):D250–D254
Bairoch A, Apweiler R (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res 28(1):45
Finn RD, Alex B, Jody C, Penelope C, Eberhardt RY, Eddy SR, Andreas H, Kirstie H, Liisa H, Jaina M (2014) Pfam: the protein families database. Nucleic Acids Res 42(D1):D222–D230
Cai L, Zheng SW, Shen YJ, Zheng GD, Liu HT, Wu ZY (2018) Complete genome sequence provides insights into the biodrying-related microbial function of Bacillus thermoamylovorans isolated from sewage sludge biodrying material. Bioresour Technol 260:141–149
Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, Kyrpides NC, Hugenholtz P (2007) CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinform 8:209. https://doi.org/10.1186/1471-2105-8-209
Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SA, Medema MH (2017) AntiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 45(W1):W36–W41
Tang YJ, Martin HG, Dehal PS, Deutschbauer AM, Llora X, Meadows A, Arkin A, Keasling JD (2009) Metabolic flux analysis of Shewanella spp. reveals evolutionary robustness in central carbon metabolism. Biotechnol Bioeng 102(4):1161–1169
Castagno L, Estrella M, Sannazzaro A, Grassano A, Ruiz O (2011) Phosphate-solubilization mechanism and in vitro plant growth promotion activity mediated by Pantoea eucalypti isolated from Lotus tenuis rhizosphere in the Salado River basin (Argentina). J Appl Microbiol 110(5):1151–1165. https://doi.org/10.1111/j.1365-2672.2011.04968.x
Bakhshandeh E, Rahimian H, Pirdashti H, Nematzadeh GA (2014) Phosphate solubilization potential and modeling of stress tolerance of rhizobacteria from rice paddy soil in northern Iran. World J Microbiol Biotechnol 30(9):2437–2447. https://doi.org/10.1007/s11274-014-1669-1
Khalid A, Arshad M, Zahir ZA (2004) Screening plant growth-promoting rhizobacteria for improving growth and yield of wheat. J Appl Microbiol 3:473–480
Delshadi S, Ebrahimi M, Shirmohammadi E (2017) Influence of plant-growth-promoting bacteria on germination, growth and nutrient uptake of Onobrychis sativa L., under drought stress. J Plant Interact 12(1):200–208
Bidabadi SS, Mehralian M (2020) Seed bio-priming to improve germination, seedling growth and essential oil yield of Dracocephalum kotschyi Boiss, an endangered medicinal plant in Iran. Gesunde Pflanzen 72:17–27. https://doi.org/10.1007/s10343-019-00478-2
Mahmood A, Turgay OC, Farooq M, Hayat R (2016) Seed biopriming with plant growth promoting rhizobacteria: a review. FEMS Microbiol Ecol 92(8):1–14. https://doi.org/10.1093/femsec/fiw112
Pan S, Rasul F, Li W, Tian H, Mo Z, Duan M, Tang X (2013) Roles of plant growth regulators on yield, grain qualities and antioxidant enzyme activities in super hybrid rice (Oryza sativa L.). Rice (NY) 6(1):9. https://doi.org/10.1186/1939-8433-6-9
Esfahani N, Mostajeran A (2011) Rhizobial strain involvement in symbiosis efficiency of chickpea-rhizobia under drought stress: plant growth, nitrogen fixation and antioxidant enzyme activities. Acta Physiol Plant 33(4):1075–1083
He J, Qin J, Long L, Ma Y, Li H, Li K, Jiang X, Liu T, Polle A, Liang Z, Luo ZB (2011) Net cadmium flux and accumulation reveal tissue-specific oxidative stress and detoxification in Populus × canescens. Physiol Plantarum 143:50–63. https://doi.org/10.1111/j.1399-3054.2011.01487.x
Bélanger M, Burrows LL, Lam JS (1999) Functional analysis of genes responsible for the synthesis of the B-band O antigen of Pseudomonas aeruginosa serotype O6 lipopolysaccharide. Microbiology (Reading, England) 145(Pt 12):3505–3521. https://doi.org/10.1099/00221287-145-12-3505
Han AW, Sandy M, Fishman B, Trindade-Silva AE, Soares CA, Distel DL, Butler A, Haygood MG (2013) Turnerbactin, a novel triscatecholate siderophore from the shipworm endosymbiont Teredinibacter turnerae T7901. PloS One 8(10):e76151. https://doi.org/10.1371/journal.pone.0076151
Arguelles-Arias A, Ongena M, Halimi B, Lara Y, Brans A, Joris B, Fickers P (2009) Bacillus amyloliquefaciens GA1 as a source of potent antibiotics and other secondary metabolites for biocontrol of plant pathogens. Microb Cell Fact 8:63
Stella M, Halimi M (2015) Gluconic acid production by bacteria to liberate phosphorus from insoluble phosphate complexes. J Trop Agric Food Sci 43:41–45
Fincheira P, Quiroz A (2018) Microbial volatiles as plant growth inducers. Microbiol Res 208:63–75
de Souza RSC, Armanhi JSL, de Damasceno N, B, Imperial J, Arruda P, (2019) Genome sequences of a plant beneficial synthetic bacterial community reveal genetic features for successful plant colonization. Front Microbiol. https://doi.org/10.3389/fmicb.2019.01779
Farías-Rodríguez R, Mellor RB, Arias C (1998) The accumulation of trehalose in nodules of several cultivars of common bean (Phaseolus vulgaris) and its correlation with resistance to drought stress. Physiol Plantarum 102(3):353–359
Suárez R, Wong A, Ramírez M, Barraza A, Orozco MDC, Cevallos MA, Lara M, Hernández G, Iturriaga G (2008) Improvement of drought tolerance and grain yield in common bean by overexpressing trehalose-6-phosphate synthase in rhizobia. Mol Plant Microb Interact 21(7):958–966
Brodmann A, Schuller A, Ludwig-Müller J, Aeschbacher R, Wiemken A, Boller T, Wingler A (2002) Induction of trehalase in Arabidopsis plants infected with the trehalose-producing pathogen Plasmodiophora brassicae. Mol Plant Microb Interact 15(7):693–700. https://doi.org/10.1094/MPMI.2002.15.7.693
Isidorov VA, Lech P, Żółciak A, Rusak M, Szczepaniak L (2008) Gas chromatographic-mass spectrometric investigation of metabolites from the needles and roots of pine seedlings at early stages of pathogenic fungi Armillaria ostoyae attack. Trees 22(4):531–542
Funding
This study was supported by the Shanghai Agriculture Applied Technology Development Program (Grant No. 2020-2-1), the National Natural Science Foundation of China (31801511), the Shanghai Agricultural Science and Technology Innovation Action Plan No. 21N31900800, the Shanghai Natural Science Foundation (No. 19ZR1436800), the Shanghai Sailing Program (No. 20YF1443000), the Technology Support Project (No. KJZC202008), the Shanghai Fresh Corn Technology System Project (No.10 2017), and the SAAS Program for Excellent Research Team (No. 2022 (016)) for financial support to this study.
Author information
Authors and Affiliations
Contributions
SSW, JBW, and YFZ analyzed the genomic data and were responsible for experiment. SSW and JBW wrote the manuscript. SSW, JBW, and YNH edited the manuscript. XMT conceived the experimental design and edited the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, S., Wang, J., Zhou, Y. et al. Isolation, Classification, and Growth-Promoting Effects of Pantoea sp. YSD J2 from the Aboveground Leaves of Cyperus Esculentus L. var. sativus. Curr Microbiol 79, 66 (2022). https://doi.org/10.1007/s00284-021-02755-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-021-02755-8