Abstract
Plant growth-promoting bacteria (PGPB) as biofertilizer plays an important role in agriculture practices. In this study, we isolated and identified plant-associated bacteria Leclercia adecarboxylata (kcgeb_e1) from the root region of the halophytic plant Sesuvium verrucosum. We tested its physiological activity and the effect of inoculation, with and without salt, on photosynthesis using Cajanus cajan. Further, we sequenced the whole genome of L. adecarboxylata (kcgeb_e1) and carried out pangenome analysis with 12 other genomes of the same species, which highlights unique genes enriched for pathways involved in abiotic stress tolerance (salinity, drought and heat) and carbohydrate transport. Moreover, gene families involved in abiotic stress tolerance, host adhesion, and transport were under positive selection (e.g., Aldo/keto reductase family, Hemagglutinin, Porin, and sugar transport). We observed a loss of ACC deaminase gene in this pangenome; however, this strain can still produce 1-aminocyclopropane-1-carboxylate (ACC), an enhancer of abiotic stress, which suggests that its homologue, d-cysteine sulfatase, has a bifunctional activity. In addition, this strain has Indole acetic acid (IAA) and phosphate solubilization activity. Combining these findings with the efficiency of colonizing the root surface of Solanum lycopersicum, this strain showed remarkable enhancement of photosynthesis, comparing control to inoculated plants. This increase in photosynthesis is consistent with an increase in sucrose under salt treatment, but not in glucose and fructose, which acts as a sensor in opposing the negative effect of salinity and promoting sustainable growth. Given all this, our study suggests that this PGPB can act as a biofertilizer for sustainable agriculture.
Article Highlights
-
Isolation of the plant-associated bacterium L. adecarboxylata (kcgeb_e1) from the root region of the halophytic plant S. verrucosum.
-
The detailed characterization of L. adecarboxylata (kcgeb_e1) revealed its plant growth-promoting activities such as IAA, ACC and phosphate solubilization.
-
Whole genome sequencing and pan-genome analysis reveal the enrichment of genes involved in abiotic stress tolerance and carbohydrate transport pathways.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Extensive efforts have been applied to improve abiotic stress tolerance of agriculture crops via conventional breeding or genetic modification, but recently most of the attention from researchers has focused on plant-associated microbes for sustainable application in agriculture [1,2,3,4]. Sesuvium verrucosum (Aizoaceae; subfamily of Sesuvioideae) is a drought- and salt-tolerant perennial plant, found in the Arabian Peninsula as well as some parts of the tropical and subtropical regions of the Americas [5]. With its natural ecological plasticity, and abiotic stress tolerance, S. verrucosum serves quite well to mine plant-associated microbes with potential for sustainable agriculture.
Bacteria that engage in a beneficial relationship with plants are termed plant growth-promoting bacteria (PGPB). To qualify as a PGPB, a positive improvement of the plant growth and fitness would be the key upon inoculation [6]. PGPB enhances plant growth by nitrogen fixation, phosphorous solubilization, activation of plant phytohormones cascades as well as antibiotics and siderophores productions [7]. For instance, rhizobacteria use tryptophan and other molecules from root exudates to convert to indole-3-acetic acid (IAA, IAA3), which triggers the plant’s endogenous auxin signaling pathway involved in the differentiation and proliferation of plant cells [8]. The accumulation of IAA induces the transcription of ACC synthase genes that increase the ACC concentration, which in return increases the harmful effect of ethylene levels in the plant [8]. Under stressful environmental conditions, PGPB with ACC deaminase activity can break down the excess of ACC, and further reduces ethylene levels [9]. However, the effect of these naturally occurring rhizobacteria from halophytic plants under different environmental stressors on commercial crops is not well understood.
Environmental stresses, such as salinity, drought, and heat affect crop productivity via impairment of electron transport, which leads to the accumulation of reactive oxygen radicals (H2O2, O2− and OH−) that end up damaging the photosynthetic apparatus [10]. As a primary source of carbon and energy in cells, sugars are synthetized during photosynthesis. The sugars are assimilated and transported from the source to different tissues through the carbohydrate partitioning process [11]. During the photosynthetic reaction, sucrose is the main product produced in the cytosol and transported to other sink organs [12], where it acts as signaling molecules to promote growth and differentiation [13]. Sugars as a source of carbon skeletons are involved in the growth and differentiation process in different plants acting as intermediate metabolites, osmolytes, storage substances, and signals for abiotic and biotic stresses [14,15,16,17].
Pangenome analysis of PGPB is important to elucidate genes that impact niche specificity. Extensive pangenomes are known to be the outcome of adaptive evolution, shaping organism fitness [18]. By linking core and accessory genes of a pangenome of a prokaryote to its lifestyle, we will be able to mine novel genes that could confer benefits to the host, as well as fitness advantage in extreme environments under different stresses [19, 20]. Even if we discover missing genes in the plant-associated bacteria, their absence may not necessarily be harmful, as it could contribute positively to the bacteria's adaptation to changing environments [21].
High yield through conventional farming practices requires the constant use of costly and toxic commercial chemical fertilizers [22]. Therefore, a recent shift toward friendly solutions for sustainable and solely organic agriculture systems is evident [22, 23]. The use of PGPB is an attractive system that can replace the harmful effect of commercial fertilizers and supplements. Some PGPB are commercially produced to improve growth in supplying nutrients to plants to sustain healthy soil environment productivity [6, 24].
Leclercia adecarboxylata, a Gram-negative bacterium belonging to the Enterobacteriaceae family, has primarily been documented in samples obtained from humans and other animals [25, 26]. In recent years, various strains of this bacterium have been discovered in diverse agricultural fields, investigations on these strains confirm the potential plant growth-promoting activity in various plants [27,28,29,30]. In this study, we isolated plant-associated bacteria Leclercia adecarboxylata (kcgeb_e1) from the root region of the halophyte S. verrucosum and we generated a pangenome analysis of 12 of this strain, to mine novel genes involved in their adaptation and plant growth promotion traits. In addition, we tested its PGPB capability through localization in root tissue as well as IAA, ACC, and phosphorus solubilization activity. Furthermore, we examined its capability to improve the plant growth, photosynthesis performance and sugar metabolite content through artificial inoculation and further imposing plants under various salt stress conditions.
2 Materials and methods
2.1 Bacterial isolation and selection from S. verrucosum
S. verrucosum plants were randomly uprooted and collected according to their natural occurrence from the UAE University farm located at Nahshilah (140 km from Al Ain City, Abu Dhabi, UAE) in sterile plastic bags, and kept in an icebox. Root samples were washed directly with tap water to remove soil particles adhering to the roots, and immediately surface–sterilized according to the previously described protocol [31] with slight modifications. Root samples were first surface sterilized with 70% ethanol for 1 min, followed by 2.5% sodium hypochlorite (NaOCl) for 20 min, and then 70% ethanol for 30 s. Subsequently, roots were washed three times with sterile distilled water (SDW). Finally, roots were cut into small pieces (1.5–2.0 mm length) and ground with mortar and pestle into fine powder in the presence of liquid Nitrogen. 10 g of the powder was added to sterile falcon bottles containing 90 ml of SDW and incubated in a shaker (180 rpm) adjusted at 28 ºC for 72 h. Serial dilutions were prepared (up to 10–7) in SDW from the supernatant and plated onto LB medium (10 g d–glucose, 5 g yeast extract, 10 g tryptone, and 15 g agar per liter). The plates were kept in an incubator at 28 ºC for 72 h. From the plates, we observed the growth of four bacterial colonies, one of which, named kcgeb_e1, showed a positive sign as a potential PGPB, and was used for downstream physiological and genomic analysis.
2.2 Bacterial species identification and whole genome sequencing
DNA isolation, quality check, Sanger sequencing, and species identification based on 16 s rRNA gene were carried out according to our previous article A 16 s rDNA based dendrogram was created by NJ method using MEGA X tool [32] with the bootstrap support of 1000. For the whole genome assembly, we generated both long (Oxford Nanopore MinION) and short (Illumina) reads. MinION long reads were generated according to the procedure described in the article [33]. Illumina shot-gun read library preparation was carried out by NEBNext® Ultra™ II DNA library preparation kit, and the sequencing was performed on Illumina NovaSeq 6000 sequencer (150 bp paired-end chemistry).
The FAST5 data generated from the MinION sequencer were base called, demultiplexed, and adapter trimmed using Guppy v.3.3.2 (implemented in MinKNOW v.3.5.5 interface, Oxford Nanopore, Cambridge UK). The long-read error correction, trimming, and length filtration (read length > 1000 bp) were carried out using CANU v.1.8 tool [34] and corrected reads were used for genome assembly. Illumina raw data quality was confirmed with FastQC [35] tool. Adapter, ambiguous bases, and low-quality regions found in the Illumina reads were trimmed using Trimmomatic v.0.39 program [36]. By using both Illumina and MinION reads, hybrid de novo genome assembly was performed using Unicycler v0.4.8 [37] with default settings (including genome error correction, genome circulation, and genome rotation). Assessment of the quality and completeness of the assembled genome was performed using BUSCO v.4.1.4 [38]. Gene prediction and genome annotation were carried out using NCBI-PGAP [39] and Prokka [40] pipeline.
2.3 Indole-3-acetic acid (IAA) production and quantification
To analyze the IAA production, the isolated strain (kcgeb_e1) was grown in Dworkin and Foster (DF) salt minimal medium supplemented with 2 mg/ml l-tryptophan (Sigma-Aldrich Co. USA) and kept at 28 ºC in a shaking incubator (200 rpm). A 2 ml of cell culture supernatant was collected after 48 h of cultivation by centrifugation. The IAA concentration in the cell culture supernatant was measured using the colorimetric technique as described by Gordon and Weber [41] and a standard curve of IAA was generated. However, the cell culture supernatant containing IAA was mixed with Salkowski’s reagent (2:1) and incubated at room temperature for 30 min in the dark. The development of red color indicates the presence of the phytohormone IAA in the sample.
2.4 Assessment of ACC deaminase activity
The strain kcgeb_e1 was screened for ACC deaminase activity on the sterile minimal DF [42] salts media (DF salts per liter: 4.0 g KH2PO4, 6.0 g Na2HPO4, 0.2 g MgSO4.7H2O, 2.0 g glucose, 2.0 g gluconic acid, and 2.0 g citric acid with trace elements: 1 mg FeSO4.7H2O, 10 mg H3BO3, 11.19 mg MnSO4.H2O, 124.6 mg ZnSO4.7H2O, 78.22 mg CuSO4. 5 H2O, 10 mg MoO3, pH 7.2) amended with 3 mM ACC instead of (NH4)2SO4 as a sole nitrogen source [42, 43]. The inoculated plates were incubated at 28ºC for 5 days and growth was monitored daily.
2.5 Phosphate solubilization activity
The kcgeb_e1 strain was spot inoculated onto Pikovaskya’s agar medium (glucose 10.0 g, Ca2(PO4)2, 5.0 g, NaCl 0.2 g, (NH4)2SO4 0.5 g, MgSO4 0.1 g, KCl 0.2 g, Yeast extract 0.5 g, MnSO4 0.002 g, FeSO4 0.002 g, distilled water 1000 ml, Agar 20.0 g), and 5 ml of bromophenol 5% was added before pouring the plates, then the inoculated plates were incubated at 28 ºC for 5 days. After 4 to 5 days, we looked for colonies with clear phosphate-solubilizing zone.
2.6 L. adecarboxylata in vitro assay in response to salinity
To evaluate salt tolerance activity of L. adecarboxylata (kcgeb_e1), the growth of the bacteria was evaluated through optical density readings at 600 nm after incubation at 37 ºC for 72 h in LB medium, supplemented with 2–10% NaCl. Moreover, the bacterial suspension was used to inoculate sterilized conical flask with nutrient broth medium supplemented with 2–10% NaCl and incubated in a shaker (250 rpm) for 72 h. Control was used with only nutrient broth medium inoculated with the same strain. Then optical density was measured at 600 nm for all with NaCl concentrations compared to the control. Moreover, the same isolate (kcgeb_e1) was grown on starch nutrient medium supplemented with 4 and 8% NaCl and incubated at 28 ºC for 72 h.
2.7 Effect of inoculating L. adecarboxylata (kcgeb_e1) on the growth of C. cajan under salt stress.
Pigeon pea seeds (C. cajan) were surface sterilized by immersion in 70% (v/v) ethanol and 0.3% (v/v) Tween 80 for 5 min and later in a solution containing 3% (w/v) HCl and 0.3% (v/v) Tween 80 for 20 min. Seeds were washed three times with SDW. Soil for planting was sterilized at 121 ºC, 1.2 atm for 15 min in an autoclave and transferred to planting pots (20 cm diameter × 15 cm height). Four treatments with four replications were studied, each one including three pots with three seeds per pot. After seed germination, a bacterial suspension of L. adecarboxylata (kcgeb_e1) with a concentration of 106 Colony-forming units (CFU) was added at a rate of 30 ml /pot every 10 days, while for control pots only SDW was added. The treatments were as follows: C (T.0) = Pigeon pea plant without the bacterium (Control); T.1. = Pigeon pea plant with only bacterium suspension; T.2. = Pigeon pea plant with the bacterium added first and followed by adding 2% NaCl weekly, T.3. = Pigeon pea plant with 2% NaCl added first and followed by the bacterium suspension.
After 21 days, the experiment for the different treatments was stopped and leaf material from each treatment was distributed in an aluminum foil and placed in the oven at 65 °C until the weight became constant. The leaves were completely dried after 24 h. Using mortar and pestle, leaves were ground and sieved. The fine powder was stored in labeled tubes and kept in the desiccator. Further, analysis of chlorophyll and soluble sugars was performed (the detailed method in Additional file 1: Methods).
2.8 Localization of L. adecarboxylata (kcgeb_e1) in roots of S. lycopersicum.
To investigate if kcgeb_e1 is localized in the roots, green fluorescent protein (GFP) tagging of the strain was done following the method used by Chung et al. [44] for the transformation of E. coli, with slight modifications. The bacterial expression vector pZE27GFP, which is constitutively expresses GFP, was used and obtained from Addgene (MA, USA). The bacterial strain kcgeb_e1 was made competent by the protocol described by Chung et al. [44], and transformed with the plasmid pZE27GFP by heat shock method. The transformed colonies were selected based on overnight growth in LB medium supplemented with 50 μg/ml kanamycin. The presence of the plasmid in bacterial cells was confirmed by PCR with primers specific for the GFP gene (BAC-GFPF; 5′-CTACCTGTTCCATGGCCAAC-3′ and BAC-GFPR; 5′-GCTCATCCATGCCATGTGTA-3′).
Tomato seeds (S. lycopersicum) were grown and later inoculated with GFP tagged kcgeb_e1 for one month. To assess colonization of root tissue, roots from both inoculated and non-inoculated plants (control) were collected and washed thoroughly under tap water. Finally, thin sections (5–10 μm) of root were prepared with a fully automatic microtome (Acculab, Canada) from control compared to inoculated samples. Sections were observed under UV light using a Leica microscope supported with (Thunder Computational Clearing) for the image (the detailed method in Additional file 1: Methods).
2.8.1 Pangenome analysis of L. adecarboxylata genomes
Prokka gene annotation for L. adecarboxylata (kcgeb_e1), as well as the other 12 genomes from NCBI (https://www.ncbi.nlm.nih.gov) were loaded in Anvi’o [45]. COG annotation was run in Anvi’o using BlastP. Pangenome analysis using Anvi’o was run with default parameters, except for the homology-based search done with BlastP instead of diamond, when comparative protein sequences were performed for clustering. Total genes, unique and core genes, and the average for the pangenomes clusters were plotted using R [46].
2.8.2 Detection of gene families under selection
Fustr pipeline [47] was used to detect if there were any gene families within L. adecarboxylata genomes under selection. We used our annotated strain transcripts and each transcript for the other 12 strains annotated and published on NCBI (https://www.ncbi.nlm.nih.gov). First, the pipeline translates sequences using TransDecoder [48], predict open reading frames, infer homology using BlastP [49], clustering with SiLiX [50], generation of multiple alignment using MAFFT [51] and building a phylogenetic tree for each gene family using FastTree [52]. After that, selection analysis is carried out with gene families with at least 15 members using a site-specific test for positive selection using the tool codeml in PAML [53], and the log-likelihood was compared to models under neutrality. A list of gene families under selection highlighting the number of sites is generated, where the ratio of non-synonymous to synonymous sites exceeds 1, reflecting a strong positive selection.
3 Results
3.1 L. adecarboxylata genome sequencing and annotation
The present work describes the isolation, sequencing and functional analysis of L. adecarboxylata (kcgeb_e1), isolated from S. verrucosum roots from a farm in the United Arab Emirates. Sanger sequencing of a ~ 1.5 kb 16S rRNA gene amplicon showed no evidence of cross-contamination and revealed high similarity with L. adecarboxylata. The phylogenetic relationship between the sequenced strain and other bacteria is shown in Fig. 1A. A whole genome assembly of this strain was created using both Oxford Nanopore and Illumina technologies. In total, 528,303 MinION long reads (read length range: 53 to 107,230 bp) which represents 874,810,700 total bp of sequence (~ 180X coverage; N50 of 3347 bp) and 9,974,177 Illumina (paired-end) PE short reads (150 bp chemistry) were generated for this study. Further, Illumina were quality-trimmed which resulted in, 9,927,476 PE reads (reads length range: 50 to 150 bp). The hybrid genome assembly generated a complete single circular genome (size 4,695,432 bp and GC% ~ 56.43) and a circular plasmid (size 42,288 bp and GC% ~ 50.78) (Fig. 1B, C). Together, the 16S rRNA gene sequence and assembled whole genome of the isolated bacterial strain confirmed it to be L. adecarboxylata. A BUSCO analysis of the assembled genome (using bacteria_odb10) resulted in 99.2% single copy complete BUSCOs and 0.8% missing BUSCOs. Genome annotation resulted in 4496 gene models (CDSs: 4337, rRNA: 25, tRNA: 84, ncRNA: 11 and pseudo genes: 39).
3.2 IAA, ACC, phosphorus activity relevant to abiotic stress tolerance
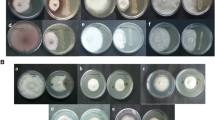
The results demonstrated that L. adecarboxylata has high IAA activity as indicated by the formation of dark red color when tryptophan was used as substrate in a colorimetric assay (Fig. 2A). The average optical density for 3 replicates was 0.948 ± 0.061, and the concentration of IAA was determined using a standard curve of pure IAA (Sigma) ranging between 20 and 120 µg/ml (Fig. 2B). For ACC deaminase activity, the typical morphology of the colonies incubated with ACC as substrate revealed that this strain has immense ACC activity (Fig. 2C). Moreover, the development of a clear zone around the colonies on the agar plates was considered as a positive for phosphate solubilizers (Fig. 2D).
A Indole-3-Acetic Acid (IAA) produced by endophytic bacterial strains from right (L. adecaboxylata (kcgeb_e1)) isolated from S. verrucosum roots as well as other strains isolated in the lab for the comparative purpose. B Estimate of the concentration of IAA with a standard curve of pure indole -3- acetic acid (IAA, Sigma) ranging between 20 and 120 µg/ml. C ACC deaminase produces by L. adecaboxylata (kcgeb_e1) on (Right), while NH4SO4 was used as a positive control (Left). D Phosphorous solubilization activity of the rhizobacteria L. adecaboxylata (kcgeb_e1), highlighting the hallow zone around the bacterium colony
3.3 Salinity stress tolerance of L. adecarboxylata (kcgeb_e1)
An experiment to assess growth at varying concentrations of NaCl suggests that L. adecarboxylata is salt tolerant. This strain has high salt tolerance up to 6% NaCl, but growth is attenuated at higher concentrations (Fig. 3A). Growth on starch agar plates supplemented with NaCl also suggested that growth of L. adecarboxylata is inhibited at higher salt concentrations. Colonies of this strain grown on starch agar plates supplemented with 4% and 8% NaCl, showed reduced growth at 8% NaCl (Fig. 3B).
A Optical Density for Bacterial cell Suspension (L. adecarboxylata (kcgeb_e1)) at 600 nm after 72 h. of incubation under different salinity concentration. B Growth of the rhizobacteria isolated from S. Verrucosum roots (L. adecaboxylata_kcgeb_e1) on Starch Nutrient Agar (SNA) supplemented with 4% NaCl (right) and 8% NaCl (Left)
3.4 Plant growth-promoting effect of L. adecarboxylata (kcgeb_e1) on photosynthesis and soluble sugars under salt stress
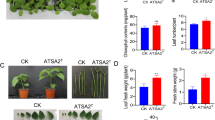
Inoculation of pigeon pea plants grown from surface-sterilized seeds with L. adecarboxylata had a growth-promoting effect in pots with three different salt treatments compared to controls. Treatment T1 had the highest level of chlorophyll a, chlorophyll b and total chlorophyll content (Total), and it had the darkest leaves compared to control and to other replicates (Fig. 4A, B, Additional file 2: Table S1). T2 and T3 treatments maintained a green foliage color with chlorophyll levels (a, b, and total) similar to each other, but their chlorophyll content was not much lower than that of treatment T1. For soluble sugars, the chromatographic profile of sugars (fructose, glucose, and sucrose) in pigeon pea leaves indicated that the three sugars were well separated, and quantification using the standard curve (Additional file 3: Fig. S1) showed a decay in the concentration of these three sugars in T1, T2, and T3 (Fig. 4C, Additional file 4: Table S2). Interestingly, the concentration of sucrose dropped in T1, but significantly increased in T2 and T3 (p < 0.05).
A Pictures of pigeon pea under control (C) and kcgeb_e1 inoculation (T1), as well as inoculated with kcgeb_e1 before (T2) and after salt treatment (T3). B Chlorophyll A, B and total concentration for the different treatments (C, T1, T2, T3) in triplicates, error bars represent standards deviation (SD). C Soluble sugars (fructose, glucose, sucrose) concentration measurements using HPCL for different treatments (C, T1, T2, T3) in triplicates, error bars represent standards deviation (SD)
3.5 Surface roots colonizing role of L. adecarboxylata (kcgeb_e1)
Microscopic examination of tomato roots, comparing those inoculated with the GPF tagged bacteria to the control, revealed colonization of rhizobacteria on the root surface, without any sign of endophytic activity (Fig. 5).
3.6 Pangenome analysis of L. adecarboxylata
Pangenome analysis of the complete genomes of 13 strains, resulted in 61,438 analyzed genes. Comparative analysis of these genomes revealed 14,490 orthologous groups and a total of 8373 unique genes (Fig. 6A, B). The pangenome consisted of individual genomes with an average of 2097 core genes, 4484 total genes, and 598 unique genes (Fig. 6A, B). Four strains (LR590464.1; CP040889.1; CP035382.1; and our strain named PROKKA_05122021[kcgeb_e1)) had more unique genes compared to other strains (Fig. 6A). kcgeb_e1 has 916 unique genes that are enriched for abiotic stress tolerance, and carbohydrate transport (Additional file 5: Table S3). Surprisingly, ACC deaminase was absent from all 13 genomes (Additional file 5: Table S3) Homologous analysis showed a match of 30% between our isolate’s d-cysteine sulfatase gene and reference ACC deaminase gene.
A Anvi’o circle plot of the 13 genomes of L. adecarboxylata. COG annotation for the different pathways is represented in bands around, and statistics about the pangenomes as histograms. B barplots of the total genes, unique and core genes in the 13 genomes of this pangenome analysis with their average counts
3.7 Gene families under positive selection
FUSTr discovered 41 families under strong positive selection in our dataset. Among these gene families, some are involved in abiotic stress tolerance (e.g., Aldo/keto reductase family, HPS70), adhesion to host cell through hemagglutination activity (e.g., Hemagglutinin repeat), and communication of solutes via transport (e.g., Porin, Major Facilitator Transport) (Additional file 6: Table S4).
4 Discussion
Plant growth-promoting microbes’ utilization is an eco-friendly practice to combat stress and several studies to date used plant growth-promoting rhizobacteria to mitigate salt stress [54]. In our study, we isolated a plant-associated bacterium, L. adecarboxylata (kcgeb_e1) from the halophyte S. verrucosum. Its potential as a PGPB activity was assessed by measuring physiological characteristics of the bacteria, and testing if it has an effect under salt stress on photosynthesis, soluble sugars—thus growth of a glycophytic crop, C. cajan. In addition, we sequenced the whole genome of this strain and carried out pangenome analysis with 12 other genomes and mined genes unique to each strain and scanned the pangenome for gene families under selection.
L. adecarboxylata is known as a pathogenic warm-blooded microbe, that belongs to the Enterobacteriaceae family [55]. It is widely known in the medical field and referred to as a human pathogen in hospital samples (urine, mucus, skin, etc.), contaminating different environments [56,57,58]. Recently another strain of L. adecarboxylata has been isolated from the rhizosphere of S. lycopersicum [27].
It has been reported that L. adecarboxylata has the capacity for hydrocarbon degradation, mineral solubilization, and, production of extracellular enzymes, and phytohormones [57, 59,60,61]. In our study, the strain showed ACC deaminase and IAA activity, which is consistent with a previous study on the same bacterium isolated from S. lycopersicum [27]. This kcgeb_e1 L. adecarboxylata rhizobacteria counteracts the negative effects of salinity on plant growth, where it secretes IAA (which promotes the growth of the plant) and a protein with ACC deaminase activity (which inhibits the negative effect of ethylene on plant growth). The cross-talking of IAA and ACC deaminase caused a decline in ethylene levels and indicated that ACC deaminase promotes plant development when IAA is present [62]. In addition, this strain thrives in the desert in the rhizosphere of this halophyte, so it must cope with nutrient deficiencies and be able to change insoluble forms of phosphorus into soluble ones for uptake by the plant, which is consistent with our results.
Pigeon peas inoculated with the strain showed significant and sustained improvement compared to the control (Fig. 4A, B). Treatments with salt before or after inoculation showed little reduction in all types of chlorophyll, compared to the bacteria inoculation by itself (Fig. 4A, B) suggesting that the strain is coping with salt stress and helping in the maintenance of photosynthesis and in return vegetative growth. As for the soluble sugars, the overall decrease was observed when comparing the inoculated treatment to control, and in salt treatment before and after inoculation. Interestingly, there was always a significant increase in glucose compared to fructose in all treatments (Fig. 4C). This finding is consistent with applications in Arabidopsis of exogenous glucose and low levels of physiologically relevant sugars, which triggers the accumulation of IAA, with its biosynthetic precursors downstream of tryptophane [63], and in return is important for protecting the plant from ethylene accumulation and salt stress. Interestingly, the observed sucrose increases in the salt inoculated strain before and after (Fig. 3C), reinforcing that sucrose is the main product of photosynthesis and explaining the sustainability of vegetative growth. Furthermore, the crosstalk between sugars and auxin signaling can explain such an increase. A recent study showed sucrose availability acts as a signal to promote auxin biosynthetic genes (TAR2) that are responsible for facilitating seed filling in pea plants [64] and a mutation in this gene will affect seed size, which was found associated with starch accumulation [65].
The pangenome is an important way to mine novel genes responsible for the fitness of an organism under different environments. The observed increase of unique genes in four strains explains that they are the products of adaptive evolution (Fig. 6B). One of them is the strain of the tomato rhizosphere, which is comparable to the environment of our strain. Two other strains were from animal excretions, a different environment, and the remaining nine strains are from human samples. The GO enrichment of the unique genes in the strain kcgeb_e1 are enriched for genes involved in abiotic stress tolerance (osmotic sensing and regulation), and this is consistent with the tolerance of this strain to 6% salt and carbohydrate transport.
Given the gene gain and loss is important to organismal fitness in different environments [18, 20], our results shows that the ACC deaminase is lost in the pangenome, which contradicts the physiological analysis which found ACC activity. The homology-based search found one homologue, d-cysteine sulfatase, that has been shown to have ACC activity that also produces ammonium as a product [66, 67]. The suggested bifunctionality of d-cysteine sulfatase implies that the loss of this gene has a fitness cost, but is rather beneficial in different environments, especially since the original pathogenic effect of this strain requires no ACC deaminase activity.
Given the adaptive and beneficial role of kcgeb_e1 in providing plant growth-promoting factors, it is important to mine gene families which are under positive selection. The FUSTr results are consistent, suggesting that selection is acting on gene families related to abiotic stress, as well as other interesting gene families involved in agglutination to cell hosts, which suggest a tight closeup relationship with the host. This is consistent with our microscopic localization of this root surface (Fig. 5), showing its close adherence to the root surface of tomato S. lycopersicum. The close adherence to the root surface also suggests interaction and transport, highlighted with genes such as porins and carbohydrates MFS.
5 Conclusion
The results of the present study suggest that kcgeb_e1 strain has great potential to sustain/ enhance vegetative growth under salinity stress, plausibly utilizing cross-talk activity of ACC and IAA, and phosphorous solubilization activity. Moreover, this strain enhanced the chlorophyll content and soluble sugar content in inoculated plants via the probable involvement of crosstalk with auxin via IAA activity. Our study suggests that this PGPB strain may be used as a biofertilizer for sustainable and ecological agricultural practices. In addition, this strain could be utilized to gain the mechanistic overview about the involvement of different biological pathways towards the abiotic stress amelioration in plant. Nevertheless, the role of cross kingdom transfer of small RNA towards the stress response is rudimentary, in this prospect this strain could be utilized to explore the role of bacterial small RNA in cross kingdom transfer to tomato plant towards abiotic resilience and further for microbial engineering to boost sustainable agriculture under harsh climatic condition.
Data availability
The Illumina and Oxford Nanopore reads synthesized during this study have been deposited in the NCBI-SRA database under the BioProject id: PRJNA842664; Biosample: SRR19415912 (Illumina PE reads) and SRR19415911 (Oxford Nanopore). The assembled genome and plasmid sequence were submitted in the NCBI-Genbank database; Accession numbers: CP098325 (genome) and CP098326 (plasmid).
Code availability
Not applicable.
References
Hou S, Thiergart T, Vannier N, Mesny F, Ziegler J, Pickel B, et al. A microbiota–root–shoot circuit favours Arabidopsis growth over defence under suboptimal light. Nature Plants. 2021;7(8):1078–92.
Bhat MA, Kumar V, Bhat MA, Wani IA, Dar FL, Farooq I, et al. Mechanistic insights of the interaction of plant growth-promoting rhizobacteria (PGPR) with plant roots toward enhancing plant productivity by alleviating salinity stress. Front Microbiol. 2020;11:1952.
de Vries FT, Griffiths RI, Knight CG, Nicolitch O, Williams A. Harnessing rhizosphere microbiomes for drought-resilient crop production. Science. 2020;368(6488):270–4.
Omae N, Tsuda K. Plant-microbiota interactions in abiotic stress environments. Mol Plant Microbe Interact. 2022;35(7):511–26.
Bohley K, Joos O, Hartmann H, Sage R, Liede-Schumann S, Kadereit G. Phylogeny of Sesuvioideae (Aizoaceae)–Biogeography, leaf anatomy and the evolution of C4 photosynthesis. Perspect Plant Eco Evolut Systemat. 2015;17(2):116–30.
Barriuso J, Ramos Solano B, Lucas JA, Lobo AP, García-Villaraco A, Gutiérrez Mañero FJ. Ecology, genetic diversity and screening strategies of plant growth promoting rhizobacteria (PGPR). Plant-Bacter Interact Strat Techniq Promote Plant Growth. 2008;15:1–17.
Saharan B, Nehra V. Plant growth promoting rhizobacteria: a critical review. Life Sci Med Res. 2011;21(1):30.
Dakora FD, Phillips DA. Root exudates as mediators of mineral acquisition in low-nutrient environments. In: Adu-Gyamfi JJ, editor. Food security in nutrient-stressed environments: exploiting plants’ genetic capabilities. Dordrecht: Springer; 2002. p. 201–13.
Gamalero E, Glick BR. Bacterial modulation of plant ethylene levels. Plant Physiol. 2015;169(1):13–22.
Heidari M, Golpayegani A. Effects of water stress and inoculation with plant growth promoting rhizobacteria (PGPR) on antioxidant status and photosynthetic pigments in basil (Ocimum basilicum L.). J Saudi Soc Agricu Sci. 2012;11(1):57–61.
Lemoine R, Camera SL, Atanassova R, Dédaldéchamp F, Allario T, Pourtau N, et al. Source-to-sink transport of sugar and regulation by environmental factors. Front Plant Sci. 2013;4:272.
Ruan Y-L. Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu Rev Plant Biol. 2014;65:33–67.
Horacio P, Martinez-Noel G. Sucrose signaling in plants: a world yet to be explored. Plant Signal Behav. 2013;8(3): e23316.
Ciereszko I. Sucrose metabolism in plant tissues under stress conditions: key enzymes, localization and function. In: Maksymiec W, editor. Compartmentation of responses to stresses in higher plants, true or false. Kerala: Transworld Research Network; 2009. p. 193–218.
Morkunas I, Ratajczak L. The role of sugar signaling in plant defense responses against fungal pathogens. Acta Physiol Plant. 2014;36:1607–19.
Lukaszuk E, Rys M, Możdżeń K, Stawoska I, Skoczowski A, Ciereszko I. Photosynthesis and sucrose metabolism in leaves of Arabidopsis thaliana aos, ein4 and rcd1 mutants as affected by wounding. Acta Physiol Plant. 2017;39:1–12.
Sami F, Yusuf M, Faizan M, Faraz A, Hayat S. Role of sugars under abiotic stress. Plant Physiol Biochem. 2016;109:54–61.
McInerney JO, McNally A, O’Connell MJ. Why prokaryotes have pangenomes. Nat Microbiol. 2017;2(4):1–5.
Vos M, Hesselman MC, Te Beek TA, van Passel MW, Eyre-Walker A. Rates of lateral gene transfer in prokaryotes: high but why? Trends Microbiol. 2015;23(10):598–605.
Livingstone PG, Morphew RM, Whitworth DE. Genome sequencing and pan-genome analysis of 23 Corallococcus spp. strains reveal unexpected diversity, with particular plasticity of predatory gene sets. Front Microbio. 2018;9:3187.
Vos M, Eyre-Walker A. Are pangenomes adaptive or not. Nat Microbio. 2017;2(12):1576.
Orhan E, Esitken A, Ercisli S, Turan M, Sahin F. Effects of plant growth promoting rhizobacteria (PGPR) on yield, growth and nutrient contents in organically growing raspberry. Sci Hortic. 2006;111(1):38–43.
Esitken A, Pirlak L, Turan M, Sahin F. Effects of floral and foliar application of plant growth promoting rhizobacteria (PGPR) on yield, growth and nutrition of sweet cherry. Sci Hortic. 2006;110(4):324–7.
O’Connell PF. Sustainable agriculture-a valid alternative. Outlook Agricu. 1992;21(1):5–12.
Ying Y, Wu F, Wu C, Jiang Y, Yin M, Zhou W, et al. Florfenicol resistance in Enterobacteriaceae and whole-genome sequence analysis of florfenicol-resistant Leclercia adecarboxylata strain R25. Int J Genom. 2019;2019:9828504.
Yin Z, Hu L, Cheng Q, Jiang X, Xu Y, Yang W, et al. First report of coexistence of three different MDR plasmids, and that of occurrence of IMP-encoding plasmid in Leclercia adecarboxylata. Front Microbiol. 2019;10:2468.
Kang S-M, Shahzad R, Bilal S, Khan AL, Park Y-G, Lee K-E, et al. Indole-3-acetic-acid and ACC deaminase producing Leclercia adecarboxylata MO1 improves Solanum lycopersicum L. growth and salinity stress tolerance by endogenous secondary metabolites regulation. BMC Microbiol. 2019;19(1):80. https://doi.org/10.1186/s12866-019-1450-6.
Kang S-M, Shahzad R, Khan MA, Hasnain Z, Lee K-E, Park H-S, et al. Ameliorative effect of indole-3-acetic acid-and siderophore-producing Leclercia adecarboxylata MO1 on cucumber plants under zinc stress. J Plant Interact. 2021;16(1):30–41.
Snak A, Vendruscolo ECG, Santos MF, Fiorini A, Mesa D. Genome sequencing and analysis of plant growth-promoting attributes from Leclercia adecarboxylata. Gene Mol Bio. 2021;44:e20200130.
Chen W, Wang Z, Xu W, Hu Y. Genome sequence of Leclercia adecarboxylata QDSM01 with multiple plant growth promoting properties. Plant Growth Regulat. 2023;44:1–15.
Araújo WL, Marcon J, Maccheroni W Jr, Van Elsas JD, Van Vuurde JW, Azevedo JL. Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl Environ Microbiol. 2002;68(10):4906–14.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–9. https://doi.org/10.1093/molbev/msy096.
Salha Y, Sudalaimuthuasari N, Kundu B, AlMaskari RS, Alkaabi AS, Hazzouri KM, et al. Complete genome sequence of Phytobacter diazotrophicus strain UAEU22, a plant growth-promoting bacterium isolated from the date palm rhizosphere. Microbio Resour Announ. 2020. https://doi.org/10.1128/mra.00499-20.
Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27(5):722–36. https://doi.org/10.1101/gr.215087.116.
Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. https://doi.org/10.1093/bioinformatics/btu170.
Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6): e1005595. https://doi.org/10.1371/journal.pcbi.1005595.
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–2. https://doi.org/10.1093/bioinformatics/btv351.
Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44(14):6614–24. https://doi.org/10.1093/nar/gkw569.
Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–9. https://doi.org/10.1093/bioinformatics/btu153.
Gordon SA, Weber RP. Colorimetric estimation of indoleacetic acid. Plant Physiol. 1951;26(1):192.
Dworkin M, Foster J. Experiments with some microorganisms which utilize ethane and hydrogen. J Bacteriol. 1958;75(5):592–603.
Penrose DM, Glick BR. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol Plant. 2003;118(1):10–5.
Chung C, Niemela SL, Miller RH. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci. 1989;86(7):2172–5.
Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, et al. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ. 2015;3: e1319.
R Core Team R. R: A language and environment for statistical computing. Vienna, Austria. 2013.
Cole TJ, Brewer MS. FUSTr: a tool to find gene families under selection in transcriptomes. PeerJ. 2018;6: e4234.
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–512.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10.
Miele V, Penel S, Duret L. Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinformat. 2011;12:1–9.
Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–66.
Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26(7):1641–50.
Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–91.
Kumar K, Amaresan N, Madhuri K. Alleviation of the adverse effect of salinity stress by inoculation of plant growth promoting rhizobacteria isolated from hot humid tropical climate. Ecol Eng. 2017;102:361–6.
Richard C. Nouvelles espèces de Enterobacteriaceae. Bull Inst Pasteur. 1984;82(3):255–77.
Tamura K, Sakazaki R, Kosako Y, Yoshizaki E. Leclercia adecarboxylata gen. nov., comb. nov., formerly known as Escherichia adecarboxylata. Curr Microbio. 1986;13:179–84.
Shahzad R, Waqas M, Khan AL, Al-Hosni K, Kang S-M, Seo C-W, et al. Indoleacetic acid production and plant growth promoting potential of bacterial endophytes isolated from rice (Oryza sativa L.) seeds. Acta Biol Hung. 2017;68(2):175–86.
Kelemu S, Fory P, Zuleta C, Ricaurte Oyola J, Rao I, Lascano C. Detecting Bacterial Endophytes in tropical Grasses of the Brachiaria genus and determining their role in improving plant growth. Afr J Biotech. 2011;10:965–76.
Sarma PM, Bhattacharya D, Krishnan S, Lal B. Degradation of polycyclic aromatic hydrocarbons by a newly discovered enteric bacterium, Leclercia adecarboxylata. Appl Environ Microbio. 2004;70(5):3163–6. https://doi.org/10.1128/AEM.70.5.3163-3166.2004.
Sun K, Liu J, Gao Y, Jin L, Gu Y, Wang W. Isolation, plant colonization potential and phenanthrene degradation performance of the endophytic bacterium Pseudomonas sp. Ph6-gfp. Sci Reports. 2014;4(1):5462. https://doi.org/10.1038/srep05462.
Yang P-X, Ma L, Chen M-H, Xi J-Q, He F, Duan C-Q, et al. Phosphate solubilizing ability and phylogenetic diversity of bacteria from P-rich soils around Dianchi lake drainage area of China. Pedosphere. 2012;22(5):707–16. https://doi.org/10.1016/S1002-0160(12)60056-3.
Glick BR. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res. 2014;169(1):30–9. https://doi.org/10.1016/j.micres.2013.09.009.
Sairanen I, Novák O, Pěnčík A, Ikeda Y, Jones B, Sandberg G, et al. Soluble carbohydrates regulate auxin biosynthesis via PIF Proteins in Arabidopsis. Plant Cell. 2012;24(12):4907–16. https://doi.org/10.1105/tpc.112.104794.
Meitzel T, Radchuk R, McAdam EL, Thormählen I, Feil R, Munz E, et al. Trehalose 6-phosphate promotes seed filling by activating auxin biosynthesis. New Phytol. 2021;229(3):1553–65. https://doi.org/10.1111/nph.16956.
McAdam EL, Meitzel T, Quittenden LJ, Davidson SE, Dalmais M, Bendahmane AI, et al. Evidence that auxin is required for normal seed size and starch synthesis in pea. New Phytol. 2017;216(1):193–204. https://doi.org/10.1111/nph.14690.
Todorovic B, Glick BR. The interconversion of ACC deaminase and D-cysteine desulfhydrase by directed mutagenesis. Planta. 2008;229(1):193–205. https://doi.org/10.1007/s00425-008-0820-3.
Singh R, Shelke G, Kumar A, Jha P. Biochemistry and genetics of ACC deaminase: a weapon to “stress ethylene” produced in plants. Front Microbio. 2015. https://doi.org/10.3389/fmicb.2015.00937.
Acknowledgements
We thank the Ministry of Presidential Affairs, United Arab Emirates for the generous funding of Khalifa Center for Genetic Engineering and Biotechnology (KCGEB). This work was supported KCGEB internal fund.
Funding
The work was supported by internal grant of Khalifa Centre for genetic engineering and biotechnology. It is dully acknowledged
Author information
Authors and Affiliations
Contributions
Conceptualization: KMH and KMAA. Supervision: KMH and KMAA. Methodology: EES, NS, FTP, MR, AKM, RSA, BK, AMA, AKE, SG, MP, KMH. Writing–original draft preparation: KMH, EES and NS. Writing–review and editing: KMH, NS and KMAA. All authors have read and agreed to the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study does not involve any human or animal participants.
Competing interests
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Additional file 1.
Quantification of total sugar and chlorophyll content.
Additional file 2: Table S1.
shows the composition of chlorophyll in pigeon pea leaves (µg/g FW) (mean ± SD).
Additional file 3: Fig. S1.
Shows clear separation of different soluble sugars as well as the calibration curve of sucrose, fructose and glucose using high performance liquid chromatography (HPLC)
Additional file 4: Table S2.
Shows the composition of soluble sugars in pigeon pea leaves (mg/g DW) (mean ± SD).
Additional file 5.
GO functional term enrichment analysis.
Additional file 6.
Pfam annotation ID of positively selected genes.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Saeed, E.E., Sudalaimuthuasari, N., Purayil, F.T. et al. The genome, pangenome, and physiological analysis of Leclercia adecarboxylata (kcgeb_e1), a plant growth-promoting bacterium. Discov Appl Sci 6, 76 (2024). https://doi.org/10.1007/s42452-024-05703-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42452-024-05703-w