Abstract
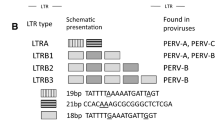
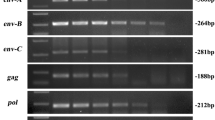
The pig appears to be the most promising animal donor of organs for use in human recipients. Among several types of pathogens found in pigs, one of the greatest problems is presented by porcine endogenous retroviruses (PERVs). Screening of the source pig herd for PERVs should include analysis of both PERV DNA and RNA. Therefore, the present study focuses on quantitative analysis of PERVs in different organs such as the skin, heart, muscle, and liver and blood of transgenic pigs generated for xenotransplantation. Transgenic pigs were developed to express the human α-galactosidase, the human α-1,2-fucosyltransferase gene, or both genetic modifications of the genome (Lipinski et al., Medycyna Wet 66:316–322, 2010; Lipinski et al., Ann Anim Sci 12:349–356, 2012; Wieczorek et al., Medycyna Wet 67:462–466, 2011). The copy numbers of PERV DNA and RNA were evaluated using real-time Q-PCR and QRT-PCR, respectively. Comparative analysis of all PERV subtypes revealed the following relationships: PERV A > PERV B > PERV C. PERV A and B were found in all samples, whereas PERV C was detected in 47 % of the tested animals. The lowest level of PERV DNA was shown in the muscles for PERV A and B and in blood samples for PERV C. The lowest level of PERV A RNA was found in the skin, whereas those of PERV B and C RNA were found in liver specimens. Quantitative analysis revealed differences in the copy number of PERV subtypes between various organs of transgenic pigs generated for xenotransplantation. Our data support the idea that careful pig selection for organ donation with low PERV copy number may limit the risk of retrovirus transmission to the human recipients.







Similar content being viewed by others
References
Bittmann I, Mihica D, Plesker R, Denner J (2012) Expression of porcine endogenous retroviruses (PERV) in different organs of a pig. Virology 433(2):329–336
Bösch S, Arnauld C, Jestin A (2000) Study of full-length porcine endogenous retrovirus genomes with envelope gene polymorphism in a specific-pathogen-free Large White swine herd. J Virol 74(18):8575–8581
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium-thiocyanate phenol-chloroform extraction. Anal Biochem 162:156–159
Chun JY, Kim KJ, Hwang IT, Kim YJ, Lee DH, Lee IK, Kim JK (2007) Dual priming oligonucleotide system for the multiplex detection of respiratory viruses and SNP genotyping of CYP2C19 gene. Nucleic Acids Res 35(6):e40
Cozzi E, Bosio E, Seveso M, Rubello D, Ancona E (2009) Xenotransplantation as a model of integrated, multidisciplinary research. Organogenesis 5(1):288–296
Cyganek-Niemiec A, Strzalka-Mrozik B, Pawlus-Lachecka L, Wszolek J, Adamska J, Kudrjavtseva J, Zhuravleva I, Kimsa M, Okla H, Kimsa M, Gudek A, Mazurek U (2012) Degradation effect of diepoxide fixation on porcine endogenous retrovirus DNA in heart valves: molecular aspects. Int J Artif Organs 35(1):25–33
Denner J, Schuurman HJ, Patience C (2009) The International Xenotransplantation Association consensus statement on conditions for undertaking clinical trials of porcine islet products in type 1 diabetes—chapter 5: strategies to prevent transmission of porcine endogenous retroviruses. Xenotransplantation 16:239–248
Dieckhoff B, Kessler B, Jobst D, Kues W, Petersen B, Pfeifer A, Kurth R, Niemann H, Wolf E, Denner J (2009) Distribution and expression of porcine endogenous retroviruses in multi-transgenic pigs generated for xenotransplantation. Xenotransplantation 16(2):64–73
Ekser B, Cooper DK (2010) Overcoming the barriers to xenotransplantation: prospects for the future. Expert Rev Clin Immunol 6(2):219–230
Food and Drug Administration (2001) PHS guideline on infectious disease issues in xenotransplantation. 1/19/2001
Gock H, Nottle M, Lew AM, d’Apice AJ, Cowan P (2011) Genetic modification of pigs for solid organ xenotransplantation. Transpl Rev (Orlando) 25(1):9–20
Harrison I, Takeuchi Y, Bartosch B, Stoye JP (2004) Determinants of high titer in recombinant porcine endogenous retroviruses. J Virol 78(24):13871–13879
Janikowska G, Strzalka B, Adamska J, Cabak B, Kowalczyk I, Mazurek U (2008) Estimation of distribution the subtypes of endogenous retroviruses PERVs in organs of domestic pigs (Sus scrofa domestica) the donors for xenotransplantations (in Polish). Farm Przegl Nauk 9–10:15–19
Kaulitz D, Mihica D, Adlhoch C, Semaan M, Denner J (2013) Improved pig donor screening including newly identified variants of porcine endogenous retrovirus-C (PERV-C). Arch Virol 158(2):341–348
Kaulitz D, Mihica D, Dorna J, Costa MR, Petersen B, Niemann H, Tönjes RR, Denner J (2011) Development of sensitive methods for detection of porcine endogenous retrovirus-C (PERV-C) in the genome of pigs. J Virol Methods 175(1):60–65
Kimsa M, Strzalka-Mrozik B, Kimsa M, Adamska J, Gola J, Lopata K, Mazurek U (2012) Quantitative estimation of porcine endogenous retrovirus release from PK15 cells. Pol J Microbiol 61:211–215
Lipinski D, Jura J, Zeyland J, Juzwa W, Maly E, Kalak R, Bochenek M, Plawski A, Szalata M, Smorag Z, Slomski R (2010) Production of transgenic pigs expressing human α-1,2-fucosyltransferase to avoid humoral xenograft rejection. Medycyna Wet 66(5):316–322
Lipinski D, Zeyland J, Plawski A, Slomski R (2012) Determination of the absolute number of transgene copies in CMVFUT transgenic pigs. Ann Anim Sci 12(3):349–356
Liu G, Li Z, Pan M, Ge M, Wang Y, Gao Y (2011) Genetic prevalence of porcine endogenous retrovirus in Chinese experimental miniature pigs. Transpl Proc 43(7):2762–2769
Löwer R, Löwer J, Kurth R (1996) The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA 93:5177–5184
Luo Y, Lin L, Bolund L, Jensen TG, Sørensen CB (2012) Genetically modified pigs for biomedical research. J Inherit Metab Dis 35(4):695–713
Machnik G, Sypniewski D, Gałka S, Loch T, Sołtysik D, Błaszczyk D, Bednarek I (2010) Changes of syncytin I expression level in HEK293 cells line after infection buy porcine endogenous retroviruses (PERV) (in Polish). Farm Przegl Nauk 12:14–20
Machnik G, Sypniewski D, Wydmuch Z, Cholewa K, Mazurek U, Wilczok T, Smorag Z, Pacha J (2005) Sequence analysis of proviral DNA of porcine endogenous retroviruses. Transpl Proc 37(10):4610–4614
Moon HJ, Park SJ, Kim HK, Ann SK, Rho S, Keum HO, Park BK (2010) Simultaneous detection and subtyping of porcine endogenous retroviruses proviral DNA using the dual priming oligonucleotide system. J Vet Sci 11(3):269–271
Prabha MS, Verghese S (2009) Polymerase chain reaction in detection of porcine endogenous retrovirus (PERV) from porcine tissues. Indian J Microbiol 49(1):68–71
Scobie L, Takeuchi Y (2009) Porcine endogenous retrovirus and other viruses in xenotransplantation. Curr Opin Organ Transpl 14(2):175–179
Semaan M, Kaulitz D, Petersen B, Niemann H, Denner J (2012) Long-term effects of PERV-specific RNA interference in transgenic pigs. Xenotransplantation 19(2):112–121
Strzalka-Mrozik B, Stanik-Walentek A, Kapral M, Kowalczyk M, Adamska J, Gola J, Mazurek U (2010) Differential expression of transforming growth factor-β isoforms in bullous keratopathy corneas. Mol Vis 16:161–166
Sypniewski D, Machnik G, Mazurek U, Wilczok T, Smorąg Z, Jura J, Gajda B (2005) Distribution of porcine endogenous retroviruses (PERVs) DNA in organs of a domestic pigs. Ann Transpl 10(2):46–51
Takeuchi Y, Patience C, Magre S, Weiss RA, Banerjee PT, Le Tissier P, Stoye JP (1998) Host range and interference studies of three classes of pig endogenous retrovirus. J Virol 72(12):9986–9991
Wieczorek J, Słomski R, Kowalski W, Smorąg Z (2011) Effect of genetic modification on the health status of transgenic pigs produced with the human α-1,2-fucosyltransferase gene. Medycyna Wet 67:462–466
Wilson CA, Wong S, VanBrocklin M, Federspiel MJ (2000) Extended analysis of the in vitro tropism of porcine endogenous retrovirus. J Virol 74(1):49–56
Zhang P, Yu P, Wang W, Zhang L, Li S, Bu H (2010) An effective method for the quantitative detection of porcine endogenous retrovirus in pig tissues. In Vitro Cell Dev Biol Anim 46(5):408–410
Acknowledgments
This study was supported by the project no. NR 12 0036 06, which was financed from 2009 to 2013 by the National Centre for Research and Development in Poland.
Conflict of interest
The authors declare that there are no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mazurek, U., Kimsa, M.C., Strzalka-Mrozik, B. et al. Quantitative Analysis of Porcine Endogenous Retroviruses in Different Organs of Transgenic Pigs Generated for Xenotransplantation. Curr Microbiol 67, 505–514 (2013). https://doi.org/10.1007/s00284-013-0397-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-013-0397-3