Abstract
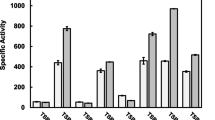
The gene (Bhbgl) encoding a β-glucosidase from the alkalophilic bacterium Bacillus halodurans C-125 was synthesized chemically via the PCR-based two-step DNA synthesis (PTDS) method and expressed in Escherichia coli. Bhbgl contained an open reading frame (ORF) of 1359 bp encoding a 453-amino acid protein belonging to glycoside hydrolase family 1 (GHF1), and the deduced molecular mass of recombinant Bhbgl (52,488 Da) was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The enzyme exhibited a high specific activity with o-nitrophenyl-β-d-glucopyranoside (oNPGlu) and an apparent K m value of 0.32 mM. With oNPGlu as the substrate, Bhbgl displayed pH and temperature optima of ~7.0 and 50°C, respectively. The enzyme was relatively stable under alkaline conditions and >50% activity was retained after incubation at pH 9.5 for 24 h at 4°C. Recombinant Bhbgl activity was inhibited by 5 mM Zn2+, Fe3+, or Cd2+, but was enhanced by 1 mM Mg2+ and other metal ions. Enzyme activity was also stimulated by at least four sugars (sucrose, d-galactose, xylose, glucose) at concentrations ranging from 50 to 800 mM.







Similar content being viewed by others
References
Hakulinen N, Paavilainen S, Korpela T, Rouvinen J (2000) The crystal structure of β-glucosidase from Bacillus circulans sp. alkalophilus: ability to form long polymeric assemblies. J Struct Biol 129:69–79
Henrissat B (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 280:309–316
Shipkowski S, Brenchley JE (2005) Characterization of an unusual cold-active β-glucosidase belonging to family 3 of the glycoside hydrolases from the psychrophilic isolate Paenibacillus sp. strain C7. Appl Environ Microbiol 17:4225–4232
Faure D (2002) The family 3 glycoside hydrolases: from housekeeping functions to host–microbe interactions. Appl Environ Microbiol 68:1485–1490
Morrissey JP, Wubben JP, Osbourn AE (2000) Stagonospora avenae secretes multiple enzymes that hydrolyze oat leaf saponins. Mol Plant Microbe Interact 13:1041–1052
Osbourn A, Bowyer P, Lunness P, Clarke B, Daniels M (1995) Fungal pathogens of oat roots and tomato leaves employ closely related enzymes to detoxify different host plant saponins. Mol Plant Microbe Interact 8:971–978
Bhat M, Bhat TS (1997) Cellulose degrading enzymes and their potential industrial applications. Biotechnol Adv 15:583–620
Bhatia Y, Mishra S, Bisaria VS (2002) Microbial β-glucosidases: cloning, properties, and applications. Crit Rev Biotechnol 22:375–407
Hu Y, Luan HW, Zhou K, Ge GB, Yang SL, Yang L (2009) Purification and characterization of a novel glycosidase from the china white jade snail (Achatina fulica) showing transglycosylation activity. Enzyme Microb Technol 43:35–42
Han YJ, Chen HZ (2008) Characterization of β-glucosidase from corn stover and its application in simultaneous saccharification and fermentation. Bioresource Technol 99:6081–6087
Sue M, Ishihara A, Iwamura H (2000) Purification and characterization of a hydroxamic acid glucoside β-glucosidase from wheat (Triticum aestivum L.) seedlings. Planta 210:432–438
Jiang C, Ma G, Li S, Hu T, Che Z, Shen P, Yan B (2009) Characterization of a novel β-glucosidase-like activity from a soil metagenome. J Microbiol 47:542–548
Benoliel B, Torres MJ, Moraes LM (2010) Expression of a glucose-tolerant β-glucosidase from Humicola grisea var. thermoidea in Saccharomyces cerevisiae. Appl Biochem Biotechnol 9:8732–8737
Hong J, Tamaki H, Kumagai H (2007) Cloning and functional expression of thermostable β-glucosidase gene from Thermoascus aurantiacus. Appl Microbiol Biotechnol 73:1331–1339
Hong MR, Kim YS, Park CS, Lee JK, Oh DK (2009) Characterization of a recombinant β-glucosidase from the thermophilic bacterium Caldicellulosiruptor saccharolyticus. J Biosci Bioeng 108:36–40
Takami H, Nakasone K, Takaki Y, Maeno G, Sasaki R, Masui N, Fuji F (2000) Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res 28:4317–4331
Ikura Y, Horikoshi K (1979) Isolation and some properties of β-galactosidase producing bacteria. Agric Biol Chem 43:85–88
Honda H, Kudo T, Horikoshi K (1985) Molecular cloning and expression of the xylanase gene of alkalophilic Bacillus sp. strain C-125 in Escherichia coli. J Bacteriol 161:784–785
Honda Y, Kitaoka M (2004) A family 8 glycoside hydrolase from Bacillus halodurans C-125 (BH2105) is a reducing end xylose-releasing exo-oligoxylanase. J Biol Chem 279:55097–55103
Smaali I, Remond C, O’Donohue MJ (2006) Expression in Escherichia coli and characterization of β-xylosidases GH39 and GH-43 from Bacillus halodurans C-125. Appl Microbiol Biotechnol 73:582–590
Navarro-Fernandez J, Martinez-Martinez I, Montoro-Garcia S, Garcia-Carmona F, Takami H, Sanchez-Ferrer A (2008) Characterization of a new rhamnogalacturonan acetyl esterase from Bacillus halodurans C-125 with a new putative carbohydrate binding domain. J Bacteriol 190:1375–1382
Xiong AS, Yao QH, Peng RH, Li X, Fan HQ, Cheng ZM, Li Y (2004) A simple, rapid, high-fidelity and cost-effective PCR-based two-step DNA synthesis method for long gene sequences. Nucleic Acids Res 32:e98
Sambrook S, Russell D (2001) Molecular cloning: a laboratory manual, 3rd edn. CSHL Press, Woodbury, NY
Stanislawski J (1991) Enzyme kinetics, version 1.5. Trinity Software, Fort Pierce, FL
Bradford MM (1976) A rapid and sensitive method for detecting microgram amounts of protein utilizing the principle of protein–dye binding. Anal Biochem 72:248–254
Higgins D, Thompson J, Gibson T (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL (2010) The Pfam protein families database. Nucleic Acids Res 38:211–222
Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int J Neural Syst 8:581–599
Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580
Peng RH, Xiong AS, Yao QH (2006) A direct and efficient PAGE-mediated overlap extension method for gene multiple-site mutagenesis. Appl Microbiol Biotechnol 73:234–240
Xiong AS, Yao QH, Peng RH, Duan H, Li X, Fan HQ, Cheng ZM (2006) PCR-based accurate synthesis of long DNA sequences. Nat Protoc 1:791–797
Sanz-Aparicio J, Hermoso JA, Martinez-Ripoll M, Lequerica JL, Polaina J (1998) Crystal structure of β-glucosidase A from Bacillus polymyxa: insights into the catalytic activity in family 1 glycosyl hydrolases. J Mol Biol 275:491–502
Saha BC, Shelby NF, Rodney JB (1994) Production, purification, and properties of a thermostable β-glucosidase from a color variant strain of Aureobasidium pullulans. Appl Environ Microbiol 103:774–3780
Kaper T, Lebbink JHG, Pouwels J, Kopp J, Schulz GE, van der Oost J, de Vos WM (2000) Comparative structural analysis and substrate specificity engineering of the hyperthermostable β-glucosidase CelB from Pyrococcus furiosus. Biochemistry 39:4963–4970
Xiong AS, Peng RH, Cheng ZM, Li Y, Liu JG, Zhuang J, Gao F, Xu F, Qiao YS, Zhang Z, Chen JM, Yao QH (2007) Concurrent mutations in six amino acids in β-glucuronidase improve its thermostability. Protein Eng Des Sel 20:319–325
Karnchanatat A, Petsom A, Sangvanich P, Piaphukiew J, Whalley AJ, Reynolds CD, Sihanonth P (2007) Purification and biochemical characterization of an extracellular beta-glucosidase from the wood-decaying fungus Daldinia eschscholzii. FEMS Microbiol Lett 270:162–170
Lymar ES, Li B, Renganathan V (1995) Purification and characterization of a cellulose-binding β-glucosidase from cellulose-degrading cultures of Phanerochaete chrysosporium. Appl Environ Microbiol 10:2976–2980
Bhiri F, Chaabouni SE, Limam F, Ghrir R, Marzouki N (2008) Purification and biochemical characterization of extracellular β-glucosidases from the hypercellulolytic Pol6 mutant of Penicillium occitanis. Appl Biochem Biotechnol 149:169–182
Kaur J, Chadha BS, Kumar BA, Saini HS (2007) Purification and characterization of β-glucosidase from Melanocarpus sp. MTCC 3922. Electron J Biotechnol 10:260–270
Riou C, Salmon JM, Vallier MJ, Gunata Z, Barre P (1998) Purification, characterization, and substrate specificity of a novel highly glucose-tolerant β-glucosidase from Aspergillus oryzae. Appl Environ Microbiol 103:607–3614
Souza FHM, Nascimento CV, Rosa JC, Masui DC, Leone FA, Jorge JA, Furriel RPM (2010) Purification and biochemical characterization of a mycelial glucose- and xylose-stimulated β-glucosidase from the thermophilic fungus Humicola insolens. Process Biochem 45:272–278
Acknowledgments
This research was supported by the Youth Fund of the Shanghai Academy of Agricultural Sciences (Grant No. 2008-6, 2009-19), the Shanghai Basic Research Project (Grant No. 08JC1418000), the Key Project Fund of the Shanghai Municipal Committee of Agriculture (Grant No. 2008-7-5), and the Key Project Fund of the Shanghai Municipal Committee of Agriculture (Grant No. 2009-6-4). We thank Dr John Buswell for linguistic revision of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Xu, H., Xiong, AS., Zhao, W. et al. Characterization of a Glucose-, Xylose-, Sucrose-, and d-Galactose-Stimulated β-Glucosidase from the Alkalophilic Bacterium Bacillus halodurans C-125. Curr Microbiol 62, 833–839 (2011). https://doi.org/10.1007/s00284-010-9766-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-010-9766-3