Abstract
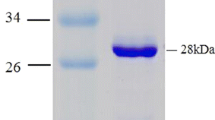
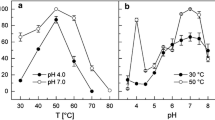
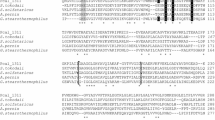
A 4-hydroxyphenylacetic acid (4-HPA) hydroxylase-encoding gene, on a 2.7-kb genomic DNA fragment, was cloned from the thermophile Geobacillus sp. PA-9. The Geobacillus sp. PA-9 4-HPA hydroxylase gene, designated hpaH, encodes a protein of 494 amino acids with a predicted molecular mass of 56.269 Da. The deduced amino-acid sequence of the hpaH gene product displayed <30% amino-acid sequence identity with the larger monooxygenase components of the previously characterized two-component 4-HPA 3-hydroxylases from Escherichia coli W and Klebsiella pneumoniae M5a1. A second oxidoreductase component was not present on the 2.7-kb genomic DNA fragment. The deduced amino-acid sequence of a second C-terminal truncated open reading frame, designated hpaI, exhibited homology to extradiol oxygenases and displayed the highest amino-acid sequence identity (43%) with the 3,4-dihydroxyphenylacetate 2,3-dioxygenase of Arthrobacter globiformis, encoded by mndD. These results, along with catalytic activity observed in crude intracellular extracts prepared from Escherichia coli cells expressing hpaH, is in support of a role for hpaH in the 4-HPA degradative pathway of Geobacillus sp. PA-9.


Similar content being viewed by others
References
Ali S, Fernandez-Lafuente R, Cowan DA (1998) Meta-pathway degradation of phenolics by themophilic Bacilli. Enzyme Microbial Technol 23:462–468
Anrow EL (1937) Colorimetric determination of the components of 3,4-dihydroxyphenylalanine-tyrosine mixture. J Biol Chem 118:531–537
Bradford M (1976) A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Burlingame R, Chapman PJ (1983) Catabolism of phenylpropionic acid and its 3-hydroxy derivative by Escherichia coli. J Bacteriol 155:113–121
Buswell JA, Twomey G (1975) Utilization of phenol and cresols by Bacillus stearothermophilus, strain PH24. J Gen Microbiol 87:377–379
Chung TC, Miller HR (1993) Preparation and storage of competent Escherichia coli cells. Methods Enzymol 218:621–627
Covès J, Nivière V, Eschenbrenner M, Fontecave M (1993) NADPH-sulfite reductase from Escherichia coli. A flavin reductase participating in the generation of the free radical of the ribonucleotide reductase. J Biol Chem 268:18604–18609
Crawford DL, Barder MJ, Pometto AL, Crawford RL (1982) Chemistry of softwood lignin degradation by Streptomyces viridosporus. Arch Microbiol 131:140–145
Díaz E, Ferrández A, Prieto MA, García JL (2001) Biodegradation of aromatic compounds by Escherichia coli. Microbiol Mol Biol Rev 65:523–569
Duffner FM, Kirchner U, Bauer MP, Müller R (2000) Phenol/cresol degradation by the thermophilic Bacillus thermoglucosidasius A7: Cloning and sequence analysis of five genes involved in the pathway. Gene 256:215–221
Duffner FM, Müller R (1998) A novel phenol hydroxylase and catechol 2,3-dioxygenase from the thermophilic Bacillus thermoleovorans strain A2: Nucleotide sequence and analysis of the genes. FEMS Microbiol Lett 161:37–45
Galán B, Díaz E, Prieto MA, García JL (2000) Functional analysis of the small component of the 4-hydroxyphenylacetate 3-monooxygenase of Escherichia coli W: A prototype of a new flavin:NAD(P)H reductase subfamily. J Bacteriol 182:627–636
Gibello A, Ferrer E, Sanz J, Martín M (1995) Polymer production by a K. pneumoniae 4-HPA hydroxylase gene cloned in E. coli. Appl Environ Microbiol 61:4167–4171
Gibello A, Suárez M, Allende JL, Martín M (1997) Molecular cloning and analysis of the genes encoding the 4-hydroxyphenylacetate hydroxylase from Klebsiella pneumoniae. Arch Microbiol 167:160–166
Gurujeyalakshmi G, Oriel P (1989) Isolation of phenol-degrading Bacillus stearothermophilus and partial characterization of phenol hydroxylase. Appl Environ Microbiol 55:500–502
Hawumba FJ, Theron J, Brözel SV (2002) Thermophilic protease-producing Geobacillus from Buranga hot springs in western Uganda. Curr Microbiol 45:144–150
Kim IC, Oriel PJ (1995) Characterisation of a Bacillus stearothermophilus BR219 phenol hydroxylase gene. Appl Environ Microbiol 61:1252–1256
Laemmli UK (1970) Cleavage associated with the maturation of the head of bacteriophage T4. Nature 227:680–685
Lee J-S, Wittchen K-D, Stahl C, Strey J, Meinhardt F (2001) Cloning, expression, and carbon catabolite repression of the bam gene encoding β-amylase of Bacillus megaterium DSM319. Appl Microbiol Biotechnol 56:205–211
Prieto MA, García JL (1994) Molecular characterization of phenol 4-hydroxyphenylacetate 3-hydroxylase of Escherichia coli. A two-protein component system. J Biol Chem 269:22823–22829
Prieto MA, Pérez-Aranda A, García JL (1993) Characterization of an Escherichia coli aromatic hydroxylase with a broad substrate range. J Bacteriol 175:2162–2167
Raju GS, Kamath AV, Vaidyanathan SC (1988) Purification and properties of 4-hydroxyphenylacetic acid 3-hydroxylase from Pseudomonas putida. Biochem Biophys Res Commun 154:537–543
Wilson K (1990) Preparation of genomic DNA from bacteria. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, et al. (eds) Current protocols in molecular biology. New York, NY, Wiley Interscience, pp 2.4.1–2.4.5
Xi L, Squires CH, Monticello DJ, Childs JD (1997) A flavin reductase stimulates DszA and DszC proteins of Rhodococcus erythropolis IGTS8 in vitro. Biochem Biophys Res Commun 230:73–75
Xun L, Sandvik ER (2000) Characterization of 4-hydroxyphenylacetate 3-hydroxylase (HpaB) of Escherichia coli as a reduced flavin adenine dinucleotide-utilizing monooxygenase. Appl Environ Microbiol 66:481–486
Acknowledgments
This work was supported through grants provided by the International Foundation for Science, the Council for Development of Social Science Research in Africa and by the Makerere University Staff Development Fund.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hawumba, J.F., Brözel, V.S. & Theron, J. Cloning and Characterization of a 4-Hydroxyphenylacetate 3-Hydroxylase From the Thermophile Geobacillus sp. PA-9. Curr Microbiol 55, 480–484 (2007). https://doi.org/10.1007/s00284-007-9016-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-007-9016-5