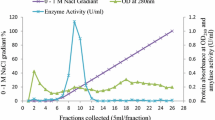
Abstract
Based on the genomic sequence and cDNA library screening, the cDNA sequence encoding an α-amylase was cloned from the filamentous white-rot fungus Phanerochaete chrysosporium and designated as pcamy1. Alignment results showed that the predicted protein has up to 43% amino acid homology to the known α-amylases in other organisms and is close to those from some filamentous fungi. Under nitrogen-starvation condition, the transcription of pcamy1 was accordingly upregulated or downregulated when soluble starch or glucose is sole carbon source. Addition of oxygen to nitrogen-limited media led to pcamy1 transcription and removal of glucose metabolic repression. The result indicated that the pcamy1 transcript was not only regulated by nutrients such as the carbon source but also by the cultivation environment, such as oxygen. This coordinate-regulatory model is likely common in P. chrysosporium. The expressed product of this gene in Escherichia coli could hydrolyze soluble starch, and its enzymatic activity was determined. As far as we know, this is the first report about cloning and expression study on the α-amylase in P. chrysosporium.







Similar content being viewed by others

Literature Cited
Belinky PA, Flikshtein N, Lechenko S, Gepstein S, Dosoretz CG (2003) Reactive oxygen species and induction of lignin peroxidase in Phanerochaete chrysosporium. Appl Envir Microbiol 69(11):6500–6506
Birch PR (1998) Targeted differential display of abundantly expressed sequences from the basidiomycete Phanerochaete chrysosporium which contain regions coding for fungal cellulose-binding domains. Curr Genet 33(1):70–76
Broda P, Birch PR, Brooks PR, Sims PF (1995) PCR-mediated analysis of lignocellulolytic gene transcription by Phanerochaete chrysosporium: Substrate-dependent differential expression within gene families. Appl Environ Microbiol 61(6):2358–2364
Caltagirone A, Weiss G, Pantopoulos K (2001) Modulation of cellular iron metabolism by hydrogen peroxide. Effects of H2O2 on the expression and function of iron-responsive element-containing mrnas in b6 fibroblasts. J Biol Chem 276(23):19738–19745
Couee I, Sulmon C, Gouesbeg G, El Amrani A (2006) Involvement of soluble sugars in reactive oxygen species balance and responses to oxidative stress in plants. J Exp Bot 57(3):449–459
Da Lage JL, Feller G, Janecek S (2004) Horizontal gene transfer from eukarya to bacteria and domain shuffling: The alpha-amylase model. Cell Mol Life Sci 61(1):97–109
Doddapaneni H, Subramanian V, Yadav JS (2005) Physiological regulation, xenobiotic induction, and heterologous expression of p450 monooxygenase gene pc-3 (cyp63a3), a new member of the cyp63 gene cluster in the white-rot fungus Phanerochaete chrysosporium. Curr Microbiol 50(6):292–298
Doodapaneni H, Yadav JS (2004) Differential regulation and xenobiotic induction of tandem p450 monooxygenase genes pc-1 (cyp63a1) and pc-2 (cyp63a2) in the white-rot fungus Phanerochaete chrysosporium. Appl Microbiol Biotechnol 65(5):559–565
Fernando T, Bumpus JA, Aust SD (1990) Biodegradation of tnt (2,4,6-trinitrotoluene) by Phanerochaete chrysosporium. Appl Environ Microbiol 56(6):1666–1671
Ferrara MA, Bon Ep, Araujo Neto JS (2002) Use of steam explosion liquor from sugar cane bagasse for liginin peroxidase production by Phanerochaete chrysosporium. Appl Biochem Biotechnol 98–100:289–300
Ganesh Kumar A, Sekaran G, Krishnamoorthy S (2006) Solid state fermentation of achras zapota lignocellulose by Phanerochaete chrysosporium. Bioresour Technol 97(13):1521–1528
Garreton V, Carpinelli J, Jordana X, Holuigue L (2002) The as-1 promoter element is an oxidative stress-responsive element and salicylic and activates it via oxidative species. Plant Physiol 130(3):1516–1526
Haseltine C, Rolfsmeier M, Blum P (1996) The glucose effect and regulation of alpha-amylase synthesis in the hyperthermophilic archaeon sulfolobus solfataricus. J Bacteriol 178(4):945–950
Hickey DA, Benkel KI, Fong Y, Benkel BF (1994) A drosophila gene promoter is subject to glucose repression in yeast cells. PNAS 91(23):11109–11112
Ho S-L, Chao Y-C, Tong W-F, Yu S-M (2001) Sugar coordinately and differentially regulates growth-and stress-related gene expression via a complex signal transduction network and multiple control mechanisms. Plant Physiol 125(2):877–890
Li D, Alic M, Brown JA, Gold MH (1995) Regulation of manganese peroxidase gene transcription by hydrogen peroxide, chemical stress, and molecular oxygen. Apply Environ Microbiol 61(1):341–345
Lu C-A, Ho T-hD, Ho S-L, Yu S-M (2002) Three novel myb proteins with one DNA binding repeat mediate sugar and hormone regulation of {alpha}-amylase gene expression. Plant Cell 14(8):1963–1980
Lu C-A, Lim E-K, Yu S-M (1998) Sugar response sequence in the promoter of a rice alpha-amylase gene serves as a transcriptional enhancer. J Biol Chem 273(17):10120–10131
Marone M, Mazzetti S, De Ritis D, Pierelli L, Scambia G (2001) Semiquantitative rt-pcr analysis to assess the expression levels of multiple transcripts from the same sample. Biol Proced Online 3:19–25
Martinez D, Larrondo LF, Putnam N, Gelpke MD, Huang K, Chapman J, et al. (2004) Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain rp78. Nat Biotechnol 22(6):695–700
Moye-Rowley WS (2003) Regulation of the transeriptional response to oxidative stress in fungi: Similarities and differences. Eukaryot Cell 2(3):381–389
Ngugen T, Sherratt PJ, Pickett CB (2003) Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol. 43:233–260
Rho D, Hodgson J, Thiboutot S, Ampleman G, Hawari J (2001) Transformation of 2,4,6-trinitrotoluene (tnt) by immobilized Phanerochaete chrysosporium under fed-batch and continuous tnt feeding conditions. Biotechnol Bioeng 73(4):271–281
Roldan-Carrillo T, Rodriguez-Varquez R, Diaz-Cervantes D, Vazquez-Torres H, Manzur-Guzman A, Torre-Dominguez A (2003) Starch-based plastic polymer degradation by the white rot fungus Phanerochaete chrysosporium grown on sugarcane bagasse pith: Enzyme production. Bioresour Technol 86(1):1–5
Sadhukhan R, Roy SK, Raha SK, Manna S, Chakrabarty SL (1992) Induction and regulation of alpha-amylase synthesis in a cellulolytic thermophilic fungus myceliophthora thermophila d14 (atcc 48104). Ind J Exp Biol 30(6):482–486
Sambrook J, Russel DW (2007) Molecular cloning: A laboratory manual, 3rd ed. New York NY: Cold Spring Harbor Laboratory Press
Schafer G, Cramer T, Suske G, Kemmner W, Wiedemann B, Hocker M (2003) Oxidative stress regulates vascular endothelial growth factor-a gene transcription through sp1-and sp3-dependent activation of two proximal gc-rich promoter elements. J Biol Chem 278(10):8190–8198
Silva A, Bacci M, Pagnocca FC, Bueno OC, Hebling MJ (2006) Starch metabolism in leucoagaricus gongylophorus, the symbiotic fungus of leaf-cutting ants. Microbiol Res 161(4):299–303
Snellinx Z, Nepovim A, Taghavi S, Vangronsveld J, Vanek T, van der Lelie D (2002) Biological remediation of explosives and related nitroaromatic compounds. Environ Sci Pollut Res Int 9(1):48–61
Storozhenko S, De Pauw P, Van Montagu M, Inze D, Kushnir S (1998) The heat-shock element is a functional component of the arabidopsis apxl gene promoter. Plant Physiol 118(3):1005–1014
Taniguchi Y, Taniguchi-Ueda Y, Mori K, Yodoi J (1996) A novel promoter sequence is involved in the oxidative stress-induced expressed of the adult t-cell leukemia-derived factor (adf)/human thioredoxin(trx) gene. Nucleic Acids Res 24(14):2746–2752
Thakkar AP, Dhamankar VS, Kapadnis BP (2006) Biocatalytic decolourisation of molasses by Phanerochaete chrysosporium. Bioresour Technol 97(12):1377–1381
Tsukamoto S, Morita S, Hirano E, Yokoi H, Masumura T, Tanaka K (2005) A novel cis-element that is responsive to oxidative stress regulates three antioxidant defense genes in rice Plant Physiol 137(1):317–327
Yoo HY, Chang MS, Rho HM (1999) The activation of the rat copper/zine superoxide dismutase gene by hydrogen peroxide through the hydrogen peroxide-responsive element and by paraquat and heat shock through the same heat shock element. J Biol Chem 274(34):23887–23892
Yoshida M, Igarashi K, Kawai R, Aida K, Samejima M (2004) Differential transcription of beta-glucosidase and cellobiose dehydrogenase genes in cellulose degradation by the basidiomycete Phanerochaete chrysosporium. FEMS Microbiol Lett 235(1):177–182
Yoshida M, Igarashi K, Wada M, Kaneko S, Suzuki N, Matsumura H (2005) Characterization of carbohydrate-binding cytochrome b562 from the white-rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol 71(8):4545–4555
Zacchi L, Burla G, Zuolong D, Harvey PJ (2000) Metabolism of cellulose by Phanerochaete chrysosporium in continuously agitated culture is associated with enhanced production of lignin peroxidase. J Biotechnol 78(2):185–192
Zhang YZ, Zylstra GJ, Oslen RH, Reddy CA (1986) Identification of cdna clones for ligmnase from Phanerochaete chrysosporium using synthetic ohgonucleotide probes. Biochem Biophys Res Commun 137(2):649–656
Acknowledgments
This work was supported by the National Science of China (Grant No. 30470984). gpd primers are courtesy of M-F. Jiang, Southwest University of Nationalities. We also thank T. Zhang at the University of Electronic Science and Technology of China for drawing three-dimensional structures.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wu, B., Hu, GK., Feng, H. et al. Cloning and Expression of an α-Amylase Gene from Phanerochaete chrysosporium . Curr Microbiol 55, 105–113 (2007). https://doi.org/10.1007/s00284-006-0600-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-006-0600-x