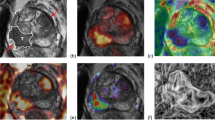
Abstract
Purpose
PI-RADS score 3 is recognized as equivocal likelihood of clinically significant prostate cancer (csPCa) occurrence. We aimed to develop a Radiomics machine learning (RML)-based redefining score to screen out csPCa in equivocal PI-RADS score 3 category.
Methods
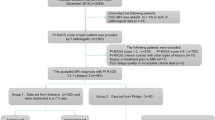
Total of 263 patients with the dominant index lesion scored PI-RADS 3 who underwent biopsy and/or follow-up formed the primary cohort. One-step RML (RML-i) model integrated radiomic features of T2WI, DWI, and ADC images all together, and two-step RML (RML-ii) model integrated the three independent radiomic signatures from T2WI (T2WIRS), DWI (DWIRS), and ADC (ADCRS) separately into a regression model. The two RML models, as well as T2WIRS, DWIRS, and ADCRS, were compared using the receiver operating characteristic-derived area under the curve (AUC), calibration plot, and decision-curve analysis (DCA). Two radiologists were asked to give a subjective binary assessment, and Cohen’s kappa statistics were calculated.
Results
A total of 59/263 (22.4%) csPCa were identified. Inter-reader agreement was moderate (Kappa = 0.435). The AUC of RML-i (0.89; 95% CI 0.88–0.90) is higher (p = 0.003) than that of RML-ii (0.87; 95% CI 0.86–0.88). The DCA demonstrated that the RML-i and RML-ii significantly improved risk prediction at threshold probabilities of csPCa at 20% to 80% compared with doing-none or doing-all by PI-RADS score 3 or stratifying by separated DWIRS, ADCRS, or T2WIRS.
Conclusion
Our RML models have the potential to predict csPCa in PI-RADS score 3 lesions, thus can inform the decision making process of biopsy.





Similar content being viewed by others
Data availability
We declared that materials described in the manuscript, including all relevant raw data, will be freely available to any scientist wishing to use them for non-commercial purposes, without breaching participant confidentiality.
References
Jambor I, Verho J. Validation of IMPROD biparametric MRI in men with clinically suspected prostate cancer: A prospective multi-institutional trial. 2019;16:e1002813.
Kumar V, Bora GS, Kumar R, Jagannathan NR. Multiparametric (mp) MRI of prostate cancer. Progress in nuclear magnetic resonance spectroscopy. 2018;105:23-40.
Weinreb JC, Barentsz JO, Choyke PL, Cornud F, Haider MA, Macura KJ, et al. PI-RADS Prostate Imaging - Reporting and Data System: 2015, Version 2. European urology. 2016;69:16-40.
Barentsz JO, Weinreb JC, Verma S, Thoeny HC, Tempany CM, Shtern F, et al. Synopsis of the PI-RADS v2 Guidelines for Multiparametric Prostate Magnetic Resonance Imaging and Recommendations for Use. European urology. 2016;69:41-9.
Rozas GQ, Saad LS, Melo H, Gabrielle HAA, Szejnfeld J. Impact of PI-RADS v2 on indication of prostate biopsy. International braz j urol : official journal of the Brazilian Society of Urology. 2019;45.
Faiena I, Salmasi A, Mendhiratta N, Markovic D, Ahuja P, Hsu W, et al. PI-RADS Version 2 Category on 3 Tesla Multiparametric Prostate Magnetic Resonance Imaging Predicts Oncologic Outcomes in Gleason 3 + 4 Prostate Cancer on Biopsy. The Journal of urology. 2019;201:91-7.
Park SY, Oh YT, Jung DC, Cho NH, Choi YD, Rha KH. Prediction of Micrometastasis (< 1 cm) to Pelvic Lymph Nodes in Prostate Cancer: Role of Preoperative MRI. AJR American journal of roentgenology. 2015;205:W328-34.
Schoots IG. MRI in early prostate cancer detection: how to manage indeterminate or equivocal PI-RADS 3 lesions? Translational andrology and urology. 2018;7:70-82.
Liddell H, Jyoti R, Haxhimolla HZ. mp-MRI Prostate Characterised PIRADS 3 Lesions are Associated with a Low Risk of Clinically Significant Prostate Cancer - A Retrospective Review of 92 Biopsied PIRADS 3 Lesions. Current urology. 2015;8:96-100.
Roethke MC, Kuru TH, Schultze S, Tichy D, Kopp-Schneider A, Fenchel M, et al. Evaluation of the ESUR PI-RADS scoring system for multiparametric MRI of the prostate with targeted MR/TRUS fusion-guided biopsy at 3.0 Tesla. European radiology. 2014;24:344-52.
Kim TJ, Lee MS, Hwang SI, Lee HJ, Hong SK. Outcomes of magnetic resonance imaging fusion-targeted biopsy of prostate imaging reporting and data system 3 lesions. World journal of urology. 2018.
Scialpi M, Aisa MC, D'Andrea A, Martorana E. Simplified Prostate Imaging Reporting and Data System for Biparametric Prostate MRI: A Proposal. AJR American journal of roentgenology. 2018;211:379-82.
Hansen NL, Koo BC, Warren AY, Kastner C, Barrett T. Sub-differentiating equivocal PI-RADS-3 lesions in multiparametric magnetic resonance imaging of the prostate to improve cancer detection. European journal of radiology. 2017;95:307-13.
Rosenkrantz AB, Meng X, Ream JM, Babb JS, Deng FM, Rusinek H, et al. Likert score 3 prostate lesions: Association between whole-lesion ADC metrics and pathologic findings at MRI/ultrasound fusion targeted biopsy. Journal of magnetic resonance imaging : JMRI. 2016;43:325-32.
Hermie I, Van Besien J, De Visschere P, Lumen N, Decaestecker K. Which clinical and radiological characteristics can predict clinically significant prostate cancer in PI-RADS 3 lesions? A retrospective study in a high-volume academic center. European journal of radiology. 2019;114:92-8.
Scialpi M, Martorana E, Aisa MC, Rondoni V, D'Andrea A, Bianchi G. Score 3 prostate lesions: a gray zone for PI-RADS v2. Turkish journal of urology. 2017;43:237-40.
Chen T, Li M, Gu Y, Zhang Y, Yang S, Wei C, et al. Prostate Cancer Differentiation and Aggressiveness: Assessment With a Radiomic-Based Model vs. PI-RADS v2. Journal of magnetic resonance imaging : JMRI. 2019;49:875-84.
Wang J, Wu CJ, Bao ML, Zhang J, Wang XN, Zhang YD. Machine learning-based analysis of MR radiomics can help to improve the diagnostic performance of PI-RADS v2 in clinically relevant prostate cancer. European radiology. 2017;27:4082-90.
Orczyk C, Villers A, Rusinek H, Lepennec V, Bazille C, Giganti F, et al. Prostate cancer heterogeneity: texture analysis score based on multiple magnetic resonance imaging sequences for detection, stratification and selection of lesions at time of biopsy. 2019;124:76-86.
Ahmed HU, Hu Y, Carter T, Arumainayagam N, Lecornet E, Freeman A, et al. Characterizing clinically significant prostate cancer using template prostate mapping biopsy. The Journal of urology. 2011;186:458-64.
Stamey TA, Freiha FS, McNeal JE, Redwine EA, Whittemore AS, Schmid HP. Localized prostate cancer. Relationship of tumor volume to clinical significance for treatment of prostate cancer. Cancer. 1993;71:933-8.
Ahmed HU, El-Shater Bosaily A, Brown LC, Gabe R, Kaplan R, Parmar MK, et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet (London, England). 2017;389:815-22.
Klein S, Staring M, Murphy K, Viergever MA, Pluim JP. elastix: a toolbox for intensity-based medical image registration. IEEE transactions on medical imaging. 2010;29:196-205.
van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer research. 2017;77:e104-e7.
Min X, Li M, Dong D, Feng Z, Zhang P, Ke Z, et al. Multi-parametric MRI-based radiomics signature for discriminating between clinically significant and insignificant prostate cancer: Cross-validation of a machine learning method. European journal of radiology. 2019;115:16-21.
Bonekamp D, Kohl S, Wiesenfarth M, Schelb P, Radtke JP, Gotz M, et al. Radiomic Machine Learning for Characterization of Prostate Lesions with MRI: Comparison to ADC Values. Radiology. 2018;289:128-37.
Ginsburg SB, Algohary A, Pahwa S, Gulani V, Ponsky L, Aronen HJ, et al. Radiomic features for prostate cancer detection on MRI differ between the transition and peripheral zones: Preliminary findings from a multi-institutional study. Journal of magnetic resonance imaging : JMRI. 2017;46:184-93.
Thestrup KC, Logager V, Baslev I, Moller JM, Hansen RH, Thomsen HS. Biparametric versus multiparametric MRI in the diagnosis of prostate cancer. Acta radiologica open. 2016;5:2058460116663046.
Kang Z, Min X, Weinreb J, Li Q, Feng Z, Wang L. Abbreviated Biparametric Versus Standard Multiparametric MRI for Diagnosis of Prostate Cancer: A Systematic Review and Meta-Analysis. AJR American journal of roentgenology. 2019;212:357-65.
Funding
This study was supported by a Key Social Development Program for the Ministry of Science and Technology of Jiangsu Province (grant number BE2017756, YDZ).
Author information
Authors and Affiliations
Contributions
HBS and YDZ contributed to conception and design and study supervision and guarantors. YH, MLB, CJW, JZ, HBS, and YDZ involved in development of methodology, acquisition of data (acquired and managed patients, provided facilities, etc.), analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis), and administrative, technical, or material support (i.e., reporting or organizing data, constructing databases). YH, YDZ, and HBS performed writing, review, and/or revision of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The author declares no conflicts of interest.
Informed consent
Informed consent was waived and all procedures performed in studies involving human participants were in accordance with the 1964 Helsinki declaration and its later amendments.
Consent for publication
This manuscript has not been published or presented elsewhere in part or in entirety and is not under consideration by another journal. The Authors agree to publication in the Journal.
Ethics approval
This study was approved by the Independent Research Ethics Boards of the First Affiliated Hospital of Nanjing Medical University (protocol 2016-SRFA-093) on Dec 2016, before data analysis was conducted.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hou, Y., Bao, ML., Wu, CJ. et al. A radiomics machine learning-based redefining score robustly identifies clinically significant prostate cancer in equivocal PI-RADS score 3 lesions. Abdom Radiol 45, 4223–4234 (2020). https://doi.org/10.1007/s00261-020-02678-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00261-020-02678-1