Abstract
Introduction
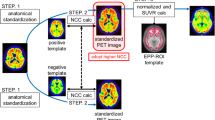
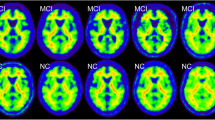
For regional quantification of nuclear brain imaging data, defining volumes of interest (VOIs) by hand is still the gold standard. As this procedure is time-consuming and operator-dependent, a variety of software tools for automated identification of neuroanatomical structures were developed. As the quality and performance of those tools are poorly investigated so far in analyzing amyloid PET data, we compared in this project four algorithms for automated VOI definition (HERMES Brass, two PMOD approaches, and FreeSurfer) against the conventional method. We systematically analyzed florbetaben brain PET and MRI data of ten patients with probable Alzheimer’s dementia (AD) and ten age-matched healthy controls (HCs) collected in a previous clinical study.
Methods
VOIs were manually defined on the data as well as through the four automated workflows. Standardized uptake value ratios (SUVRs) with the cerebellar cortex as a reference region were obtained for each VOI. SUVR comparisons between ADs and HCs were carried out using Mann-Whitney-U tests, and effect sizes (Cohen’s d) were calculated. SUVRs of automatically generated VOIs were correlated with SUVRs of conventionally derived VOIs (Pearson’s tests).
Results
The composite neocortex SUVRs obtained by manually defined VOIs were significantly higher for ADs vs. HCs (p=0.010, d=1.53). This was also the case for the four tested automated approaches which achieved effect sizes of d=1.38 to d=1.62. SUVRs of automatically generated VOIs correlated significantly with those of the hand-drawn VOIs in a number of brain regions, with regional differences in the degree of these correlations. Best overall correlation was observed in the lateral temporal VOI for all tested software tools (r=0.82 to r=0.95, p<0.001).
Conclusion
Automated VOI definition by the software tools tested has a great potential to substitute for the current standard procedure to manually define VOIs in β-amyloid PET data analysis.




Similar content being viewed by others
References
White DR, Houston AS, Sampson WF, Wilkins GP. Intra- and interoperator variations in region-of-interest drawing and their effect on the measurement of glomerular filtration rates [eng]. Clin Nucl Med. 1999;24(3):177–81.
Destrieux C, Fischl B, Dale A, Halgren E. Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. NeuroImage. 2010;53(1):1–15.
Evans AC, Marrett S, Neelin P, Collins L, Worsley K, Dai W, et al. Anatomical mapping of functional activation in stereotactic coordinate space. NeuroImage. 1992;1(1):43–53.
Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, et al. Automated Anatomical Labeling of Activations in SPM Using a Macroscopic Anatomical Parcellation of the MNI MRI Single-Subject Brain. NeuroImage. 2002;15(1):273–89.
Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain [eng]. Neuron. 2002;33(3):341–55.
Balafar MA, Ramli AR, Saripan MI, Mashohor S. Review of brain MRI image segmentation methods. Artif Intell Rev. 2010;33(3):261–74.
Ashburner J, Friston KJ. Nonlinear spatial normalization using basis functions [eng]. Hum Brain Mapp. 1999;7(4):254–66.
Collins DL, Neelin P, Peters TM, Evans AC. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space [eng]. J Comput Assist Tomogr. 1994;18(2):192–205.
Ashburner J, Friston KJ. Unified segmentation [eng]. Neuroimage. 2005;26(3):839–51.
Fischl B. Automatically parcellating the human cerebral cortex. Cereb Cortex. 2004;14(1):11–22.
Barthel H, Seibyl J, Sabri O. The role of positron emission tomography imaging in understanding Alzheimer’s disease [eng]. Expert Rev Neurother. 2015;15(4):395–406.
Barthel H, Luthardt J, Becker G, Patt M, Hammerstein E, Hartwig K, et al. Individualized quantification of brain β-amyloid burden: results of a proof of mechanism phase 0 florbetaben PET trial in patients with Alzheimer’s disease and healthy controls. Eur J Nucl Med Mol Imaging. 2011;38(9):1702–14.
Becker GA, Ichise M, Barthel H, Luthardt J, Patt M, Seese A, et al. PET Quantification of 18F-Florbetaben Binding to -Amyloid Deposits in Human Brains. J Nucl Med. 2013;54(5):723–31.
D’Agostino E, Maes F, Vandermeulen D, Suetens P. A viscous fluid model for multimodal non-rigid image registration using mutual information [eng]. Med Image Anal. 2003;7(4):565–75.
Slomka PJ, Hurwitz GA, Stephenson J, Cradduck T. Automated alignment and sizing of myocardial stress and rest scans to three-dimensional normal templates using an image registration algorithm [eng]. J Nucl Med. 1995;36(6):1115–22.
Slomka PJ, Radau P, Hurwitz GA, Dey D. Automated three-dimensional quantification of myocardial perfusion and brain SPECT [eng]. Comput Med Imaging Graph. 2001;25(2):153–64.
Radau PE, Linke R, Slomka PJ, Tatsch K. Optimization of automated quantification of 123I-IBZM uptake in the striatum applied to parkinsonism [eng]. J Nucl Med. 2000;41(2):220–7.
Radau PE, Slomka PJ, Julin P, Svensson L, Wahlund LO. Evaluation of linear registration algorithms for brain SPECT and the errors due to hypoperfusion lesions [eng]. Med Phys. 2001;28(8):1660–8.
Hammers A, Allom R, Koepp MJ, Free SL, Myers R, Lemieux L, et al. Three-dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Hum Brain Mapp. 2003;19(4):224–47.
Fischl B. FreeSurfer. NeuroImage. 2012;62(2):774–81.
Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: inflation, flattening, and a surface-based coordinate system [eng]. NeuroImage. 1999;9(2):195–207.
Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction [eng]. NeuroImage. 1999;9(2):179–94.
Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 2006;31(3):968–80.
Rowe CC, Ackerman U, Browne W, Mulligan R, Pike KL, O’Keefe G, et al. Imaging of amyloid β in Alzheimer’s disease with 18F-BAY94-9172, a novel PET tracer: proof of mechanism. Lancet Neurol. 2008;7(2):129–35.
Schain M, Varnäs K, Cselényi Z, Halldin C, Farde L, Varrone A. Evaluation of two automated methods for PET region of interest analysis [eng]. Neuroinformatics. 2014;12(4):551–62.
Aalto S, Scheinin NM, Kemppainen NM, Någren K, Kailajärvi M, Leinonen M, et al. Reproducibility of automated simplified voxel-based analysis of PET amyloid ligand [11C]PIB uptake using 30-min scanning data. Eur J Nucl Med Mol Imaging. 2009;36(10):1651–60.
Rosario BL, Weissfeld LA, Laymon CM, Mathis CA, Klunk WE, Berginc MD, et al. Inter-rater reliability of manual and automated region-of-interest delineation for PiB PET. NeuroImage. 2011;55(3):933–41.
Hutton C, Declerck J, Mintun MA, Pontecorvo MJ, Devous MD, Joshi AD. Quantification of (18)F-florbetapir PET: comparison of two analysis methods [ENG]. Eur J Nucl Med Mol Imaging. 2015;42(5):725–32.
Landau SM, Thomas BA, Thurfjell L, Schmidt M, Margolin R, Mintun M, et al. Amyloid PET imaging in Alzheimer’s disease: a comparison of three radiotracers [eng]. Eur J Nucl Med Mol Imaging. 2014;41(7):1398–407.
Edison P, Carter S, Rinne J, Gelosa G, Herholz K, Nordberg A, et al. Comparison of MRI based and PET template based approaches in the quantitative analysis of amyloid imaging with PIB-PET. NeuroImage. 2013;70:423–33.
Saint-Aubert L, Nemmi F, Péran P, Barbeau EJ, Payoux P, Chollet F, et al. Comparison between PET template-based method and MRI-based method for cortical quantification of florbetapir (AV-45) uptake in vivo [eng]. Eur J Nucl Med Mol Imaging. 2014;41(5):836–43.
Thurfjell L, Lilja J, Lundqvist R, Buckley C, Smith A, Vandenberghe R, et al. Automated quantification of 18F-flutemetamol PET activity for categorizing scans as negative or positive for brain amyloid: concordance with visual image reads [eng]. J Nucl Med : Off Publ Soc Nucl Med. 2014;55(10):1623–8.
Lundqvist R, Lilja J, Thomas BA, Lotjonen J, Villemagne VL, Rowe CC, et al. Implementation and validation of an adaptive template registration method for 18F-flutemetamol imaging data. J Nucl Med. 2013;54(8):1472–8.
Su Y, D’Angelo GM, Vlassenko AG, Zhou G, Snyder AZ, Marcus DS, et al. Quantitative Analysis of PiB-PET with FreeSurfer ROIs. PLoS ONE. 2013;8(11):e73377.
Barnes J, Foster J, Boyes R, Pepple T, Moore E, Schott J, et al. A comparison of methods for the automated calculation of volumes and atrophy rates in the hippocampus. NeuroImage. 2008;40(4):1655–71.
Pöpperl G, Radau P, Linke R, Hahn K, Tatsch K. Diagnostic performance of a 3-D automated quantification method of dopamine D2 receptor SPECT studies in the differential diagnosis of parkinsonism [eng]. Nucl Med Commun. 2005;26(1):39–43.
Koch W, Radau PE, Hamann C, Tatsch K. Clinical testing of an optimized software solution for an automated, observer-independent evaluation of dopamine transporter SPECT studies [eng]. J Nucl Med. 2005;46(7):1109–18.
Fripp J, Bourgeat P, Raniga P, Acosta O, Villemagne V, Jones G, et al. MR-less high dimensional spatial normalization of 11C PiB PET images on a population of elderly, mild cognitive impaired and Alzheimer disease patients [eng]. Med Image Comput Comput Assist Interv. 2008;11(Pt 1):442–9.
Kuhn FP, Warnock GI, Burger C, Ledermann K, Martin-Soelch C, Buck A. Comparison of PET template-based and MRI-based image processing in the quantitative analysis of C11-raclopride PET. EJNMMI Res. 2014;4(1):7.
Meyer JH, Gunn RN, Myers R, Grasby PM. Assessment of spatial normalization of PET ligand images using ligand-specific templates [eng]. NeuroImage. 1999;9(5):545–53.
Acknowledgments
We would like to thank the statistical counselling service of IMISE, University of Leipzig for their support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This project received research support by Piramal Imaging, HERMES Medical solutions, and PMOD Technologies Ltd.
Conflict of interest
OS and HB received speaker and consulting honoraria from Piramal Imaging and Siemens Healthcare. MR received travel expenses from Piramal Imaging. SH received travel grants and honoraria from General Electric (GE) Healthcare and Bayer Schering Pharma.
Ethical approval
All procedures performed involving human participants were in accordance with the ethical standards of the 1964 Helsinki declaration and its later amendments and with the standards of the institutional and national research committee. This article does not contain any studies with animals performed by any of the authors.
Informed consent
Written informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 290 kb)
Rights and permissions
About this article
Cite this article
Tuszynski, T., Rullmann, M., Luthardt, J. et al. Evaluation of software tools for automated identification of neuroanatomical structures in quantitative β-amyloid PET imaging to diagnose Alzheimer’s disease. Eur J Nucl Med Mol Imaging 43, 1077–1087 (2016). https://doi.org/10.1007/s00259-015-3300-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-015-3300-6