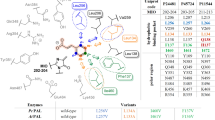
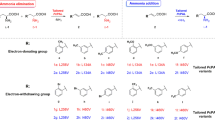
Abstract
In this study, rational design and saturation mutagenesis efforts for engineering phenylalanine ammonia-lyase from Petroselinum crispum (PcPAL) provided tailored PALs active towards challenging, highly valuable di-substituted substrates, such as the l-DOPA precursor 3,4-dimethoxy-l-phenylalanine or the 3-bromo-4-methoxy-phenylalanine. The rational design approach and saturation mutagenesis strategy unveiled identical PcPAL variants of improved activity, highlighting the limited mutational variety of the substrate specificity-modulator residues, L134, F137, I460 of PcPAL. Due to the restricted catalytic efficiency of the best performing L134A/I460V and F137V/I460V PcPAL variants, we imprinted these beneficial mutations to PALs of different origins. The variants of PALs from Arabidopsis thaliana (AtPAL) and Anabaena variabilis (AvPAL) showed higher catalytic efficiency than their PcPAL homologues. Further, the engineered PALs were also compared in terms of catalytic efficiency with a novel aromatic ammonia-lyase from Loktanella atrilutea (LaAAL), close relative of the metagenome-derived aromatic ammonia-lyase AL-11, reported recently to possess atypically high activity towards substrates with electron-donor aromatic substituents. Indeed, LaAAL outperformed the engineered Pc/At/AvPALs in the production of 3,4-dimethoxy-l-phenylalanine; however, in case of 3-bromo-4-methoxy derivatives it showed no activity, with computational results supporting the occurrence of steric hindrance. Transferring the unique array of selectivity modulator residues from LaAAL to the well-characterized PALs did not enhance their activity towards the targeted substrates. Moreover, applying the rational design strategy valid for these well-characterized PALs to LaAAL decreased its activity. These results suggest that distinct tailoring rationale is required for LaAAL/AL-11-like aromatic ammonia-lyases, which might represent a distinct PAL subclass, with natural reaction and substrate scope modified through evolutionary processes.
Key points
• PAL-activity for challenging substrates generated by protein engineering
• Rational/semi-rational protein engineering reveals constrained mutational variability
• Engineered PALs are outperformed by novel ALs of distinct catalytic site signature
Graphical abstract








Similar content being viewed by others
Data availability
The Uniprot/EMBL identifiers of all protein/nucleotide sequences used within the alignments and experimental work and the Protein Data Bank (PDB) IDs for the protein structures used within the computational part are described within the manuscript or the Supplementary Information, while the original data that support the findings of this study are included within the manuscript/supplementary information and/or are available from the corresponding author upon reasonable request.
References
Bartsch S, Bornscheuer UT (2009) A single residue influences the reaction mechanism of ammonia lyases and mutases. Angew Chem Int Ed 48(18):3362–3365. https://doi.org/10.1002/anie.200900337
Bata Z, Molnar Z, Madaras E, Molnar B, Santa-Bell E, Varga A, Leveles I, Qian RZ, Hammerschmidt F, Paizs C, Vertessy BG, Poppe L (2021) Substrate tunnel engineering aided by X-ray crystallography and functional dynamics swaps the function of MIO-enzymes. ACS Catal 11(8):4538–4549. https://doi.org/10.1021/acscatal.1c00266
de Lange B, Hyett DJ, Maas PJD, Mink D, van Assema FBJ, Sereinig N, de Vries AHM, de Vries JG (2011) Asymmetric synthesis of (s)-2-indolinecarboxylic acid by combining biocatalysis and homogeneous catalysis. ChemCatChem 3(2):289–292. https://doi.org/10.1002/cctc.201000435
Dima NA, Filip A, Bencze LC, Oláh M, Sátorhelyi P, Vértessy BG, Poppe L, Paizs C (2016) Expression and purification of recombinant phenylalanine ammonia-lyase from Petroselinum crispum. Stud Univ Babes-Bolyai Chem 61(2):21–34
Dreßen A, Hilberath T, Mackfeld U, Billmeier A, Rudat J, Pohl M (2017) Phenylalanine ammonia lyase from Arabidopsis thaliana (AtPAL2): a potent MIO-enzyme for the synthesis of non-canonical aromatic alpha-amino acids: part I: comparative characterization to the enzymes from Petroselinum crispum (PcPAL1) and Rhodosporidium toruloides (RtPAL). J Biotechnol 258:148–157. https://doi.org/10.1016/j.jbiotec.2017.04.005
Filip A, Nagy EZA, Tork SD, Bánóczi G, Toşa MI, Irimie FD, Poppe L, Paizs C, Bencze LC (2018) Tailored mutants of phenylalanine ammonia-lyase from Petroselinum crispum for the synthesis of bulky L- and D-arylalanines. ChemCatChem 10(12):2627–2633. https://doi.org/10.1002/cctc.201800258
Galman JL, Parmeggiani F, Seibt L, Birmingham WR, Turner NJ (2022) One-pot biocatalytic in vivo methylation-hydroamination of bioderived lignin monomers to generate a key precursor to L-DOPA. Angew Chem Int Ed 61(8). https://doi.org/10.1002/anie.202112855
Gloge A, Zon J, Kovari A, Poppe L, Retey J (2000) Phenylalanine ammonia-lyase: the use of its broad substrate specificity for mechanistic investigations and biocatalysis - synthesis of L-arylalanines. Chem Eur J 6(18):3386–3390. https://doi.org/10.1002/1521-3765(20000915)6:18%3c3386::Aid-chem3386%3e3.0.Co;2-5
Hardegger LA, Beney P, Bixel D, Fleury C, Gao F, Perrenoud AGG, Gu XX, Haber J, Hong T, Humair R, Kaegi A, Kibiger M, Kleinbeck F, Luu VT, Padeste L, Rampf FA, Ruch T, Schlama T, Sidler E, Udvarhelyi A, Wietfeld B, Yang Y (2020) Toward a scalable synthesis and process for EMA401, Part III: using an engineered phenylalanine ammonia lyase enzyme to synthesize a non-natural phenylalanine derivative. Org Process Res Dev 24(9):1763–1771. https://doi.org/10.1021/acs.oprd.0c00217
Kempa EE, Galman JL, Parmeggiani F, Marshall JR, Malassis J, Fontenelle CQ, Vendeville JB, Linclau B, Charnock SJ, Flitsch SL, Turner NJ, Barran PE (2021) Rapid screening of diverse biotransformations for enzyme evolution. JACS Au 1(4):508–516. https://doi.org/10.1021/jacsau.1c00027
Kille S, Acevedo-Rocha CG, Parra LP, Zhang ZG, Opperman DJ, Reetz MT, Acevedo JP (2013) Reducing codon redundancy and screening effort of combinatorial protein libraries created by saturation mutagenesis. ACS Synth Biol 2(2):83–92. https://doi.org/10.1021/sb300037w
Lovelock SL, Turner NJ (2014) Bacterial Anabaena variabilis phenylalanine ammonia lyase: a biocatalyst with broad substrate specificity. Bioorg Med Chem 22(20):5555–5557. https://doi.org/10.1016/j.bmc.2014.06.035
Mays ZJS, Mohan K, Trivedi VD, Chappell TC, Nair NU (2020) Directed evolution of Anabaena variabilis phenylalanine ammonia-lyase (PAL) identifies mutants with enhanced activities. Chem Commun 56(39):5255–5258. https://doi.org/10.1039/d0cc00783h
McKenna R, Pugh S, Thompson B, Nielsen DR (2013) Microbial production of the aromatic building-blocks (S)-styrene oxide and (R)-1,2-phenylethanediol from renewable resources. Biotechnol J 8(12):1465–1475. https://doi.org/10.1002/biot.201300035
Min K, Park K, Park DH, Yoo YJ (2015) Overview on the biotechnological production of L-DOPA. Appl Microbiol Biotechnol 99(2):575–584. https://doi.org/10.1007/s00253-014-6215-4
Moffitt MC, Louie GV, Bowman ME, Pence J, Noel JP, Moore BS (2007) Discovery of two cyanobacterial phenylalanine ammonia lyases: kinetic and structural characterization. Biochemistry 46(4):1004–1012. https://doi.org/10.1021/bi061774g
Moisa ME, Amariei DA, Nagy EZA, Szarvas N, Tosa MI, Paizs C, Bencze LC (2020) Fluorescent enzyme-coupled activity assay for phenylalanine ammonia-lyases. Sci Rep 10(1). https://doi.org/10.1038/s41598-020-75474-y
Nagy EZA, Tork SD, Lang PA, Filip A, Irimie FD, Poppe L, Toşa MI, Schofield CJ, Brem J, Paizs C, Bencze LC (2019) Mapping the hydrophobic substrate binding site of phenylalanine ammonia-lyase from Petroselinum crispum. ACS Catal 9(9):8825–8834. https://doi.org/10.1021/acscatal.9b02108
Parmeggiani F, Ahmed ST, Weise NJ, Turner NJ (2016) Telescopic one-pot condensation-hydroamination strategy for the synthesis of optically pure L-phenylalanines from benzaldehydes. Tetrahedron 72(46):7256–7262. https://doi.org/10.1016/j.tet.2015.12.063
Parmeggiani F, Weise NJ, Ahmed ST, Turner NJ (2018) Synthetic and therapeutic applications of ammonia-lyases and aminomutases. Chem Rev 118(1):73–118. https://doi.org/10.1021/acs.chemrev.6b00824
Rees DG, Jones DH (1996) Stability of L-phenylalanine ammonia-lyase in aqueous solution and as the solid state in air and organic solvents. Enzyme Microb Technol 19(4):282–288. https://doi.org/10.1016/0141-0229(95)00247-2
Reetz MT, Carballeira JD (2007) Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes. Nat Protoc 2(4):891–903. https://doi.org/10.1038/nprot.2007.72
Reetz MT, Bocola M, Carballeira JD, Zha DX, Vogel A (2005) Expanding the range of substrate acceptance of enzymes: combinatorial active-site saturation test. Angew Chem Int Ed 44(27):4192–4196. https://doi.org/10.1002/anie.200500767
Reetz MT, Carballeira DJ, Vogel A (2006) Iterative saturation mutagenesis on the basis of B factors as a strategy for increasing protein thermostability. Angew Chem Int Ed 45(46):7745–7751. https://doi.org/10.1002/anie.200602795
Reetz MT, Wang LW, Bocola M (2006b) Directed evolution of enantioselective enzymes: iterative cycles of CASTing for probing protein-sequence space. Angew Chem Int Ed 45(8):1236–1241. https://doi.org/10.1002/anie.200502746
Renard G, Guilleux JC, Bore C, Malta-Valette V, Lerner DA (1992) Synthesis of L-phenylalanine analogs by Rhodotorula glutinis. Bioconversion of cinnamic acids derivatives. Biotechnol Lett 14(8):673–678. https://doi.org/10.1007/BF01021641
Rowles I, Groenendaal B, Binay B, Malone KJ, Willies SC, Turner NJ (2016) Engineering of phenylalanine ammonia lyase from Rhodotorula graminis for the enhanced synthesis of unnatural L-amino acids. Tetrahedron 72(46):7343–7347. https://doi.org/10.1016/j.tet.2016.06.026
Sanchis J, Fernandez L, Carballeira J, Drone J, Gumulya Y, Hobenreich H, Kahakeaw D, Kille S, Lohmer R, Peyralans J, Podtetenieff J, Prasad S, Soni P, Taglieber A, Wu S, Zilly F, Reetz M (2008) Improved PCR method for the creation of saturation mutagenesis libraries in directed evolution: application to difficult-to-amplify templates. Appl Microbiol Biotechnol 81(2):387–397. https://doi.org/10.1007/s00253-008-1678-9
Stolze SC, Meltzer M, Ehrmann M (2012) Kaiser M (2012) Development of a solid-phase approach to the natural product class of Ahp-containing cyclodepsipeptides. Eur J Org Chem 8:1616–1625. https://doi.org/10.1002/ejoc.201101757
Tomoiaga RB, Tork SD, Horvath I, Filip A, Nagy LC, Bencze LC (2020) Saturation mutagenesis for phenylalanine ammonia lyases of enhanced catalytic properties. Biomolecules 10(6). https://doi.org/10.3390/biom10060838
Tork SD, Nagy EZA, Cserepes L, Bordea DM, Nagy B, Toşa MI, Paizs C, Bencze LC (2019) The production of L- and D-phenylalanines using engineered phenylalanine ammonia lyases from Petroselinum crispum. Sci Rep 9(1). https://doi.org/10.1038/s41598-019-56554-0
Tork SD, Moisa ME, Cserepes L, Filip A, Nagy LC, Irimie FD, Bencze LC (2022a) Towards a general approach for tailoring the hydrophobic binding site of phenylalanine ammonia-lyases. Sci Rep 12(1). https://doi.org/10.1038/s41598-022-14585-0
Tork SD, Nagy EZA, Tomoiagă RB, Bencze LC (2022b) Engineered, scalable production of optically pure L-phenylalanines using phenylalanine ammonia-lyase from Arabidopsis thaliana. J Org Chem. https://doi.org/10.1021/acs.joc.2c02106
Trivedi VD, Chappell TC, Krishna NB, Shetty A, Sigamani GG, Mohan K, Ramesh A, Kumar RP, Nair NU (2022) In-depth sequence-function characterization reveals multiple pathways to enhance enzymatic activity. ACS Catal 12(4):2381–2396. https://doi.org/10.1021/acscatal.1c05508
Varga A, Bata Z, Csuka P, Bordea DM, Vértessy BG, Marcovici A, Irimie FD, Poppe L, Bencze LC (2017) A novel phenylalanine ammonia-lyase from Kangiella koreensis. Stud Univ Babes-Bolyai Chem 62(3):293–308. https://doi.org/10.24193/subbchem.2017.3.25
Varga A, Csuka P, Sonesouphap O, Bánóczi G, Toşa MI, Katona G, Molnár Z, Bencze LC, Poppe L, Paizs C (2021) A novel phenylalanine ammonia-lyase from Pseudozyma antarctica for stereoselective biotransformations of unnatural amino acids. Catal Today 366:185–194. https://doi.org/10.1016/j.cattod.2020.04.002
Weise NJ, Ahmed ST, Parmeggiani F, Siirola E, Pushpanath A, Schell U, Turner NJ (2016) Intensified biocatalytic production of enantiomerically pure halophenylalanines from acrylic acids using ammonium carbamate as the ammonia source. Catal Sci Technol 6(12):4086–4089. https://doi.org/10.1039/c6cy00855k
Weise NJ, Ahmed ST, Parmeggiani F, Galman JL, Dunstan MS, Charnock SJ, Leys D, Turner NJ (2017) Zymophore identification enables the discovery of novel phenylalanine ammonia lyase enzymes. Sci Rep 7(1). https://doi.org/10.1038/s41598-017-13990-0
Yamada S, Nabe K, Izuo N (1981) Production of L-phenylalanine from trans-cinnamic acid with Rhodotorula glutinis containing L-phenylalanine ammonia-lyase activity. Appl Environ Microbiol 42(5):773–778. https://doi.org/10.1128/aem.42.5.773-778.1981
Funding
This work was financed by the Romanian Ministry of Education and Research, CNCS–UEFISCDI, project number PN-III-P1-1.1-TE-2019–2118, within PNCDI III.
Author information
Authors and Affiliations
Contributions
R.B.T. was responsible for the saturation mutagenesis experiments, preparation of whole-cell-biocatalysts, reaction monitoring by HPLC, protein purification and enzyme kinetics. S.D.T. was responsible for the mutagenesis, activity measurements of the rationally designed PALs, while A.F. contributed to primer design, sequencing interpretation and protein purification. L.C.N. was responsible for computational studies and graphical artwork. L.C.B. conceived the project, was responsible for funding, supervised all experiments, data and wrote the paper together with R.B.T. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies involving human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
253_2023_12374_MOESM1_ESM.pdf
Supplementary file1. Supplementary information of this paper consists of the mutagenesis protocol and tables of primers employed for site-specific and saturation mutagenesis, library screening conditions and the alignment of PAL/TAL/HAL/AAL of distinct origins together with their Uniprot/EMBL identifiers. Information about functional characterization of purified enzymes is also provided, including HPLC monitoring conditions, conversion, and enantiomeric excess representative chromatograms, Michaelis–Menten curves and Tm values. Detailed description of the computational work is also included. (PDF 2.59 MB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tomoiagă, R.B., Tork, S.D., Filip, A. et al. Phenylalanine ammonia-lyases: combining protein engineering and natural diversity. Appl Microbiol Biotechnol 107, 1243–1256 (2023). https://doi.org/10.1007/s00253-023-12374-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-023-12374-x