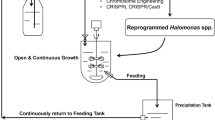
Abstract
Halomonas spp. are the well-studied platform organisms or chassis for next-generation industrial biotechnology (NGIB) due to their contamination-resistant nature combined with their fast growth property. Several Halomonas spp. have been studied regarding their genomic information and molecular engineering approaches. Halomonas spp., especially Halomonas bluephagenesis, have been engineered to produce various biopolyesters such as polyhydroxyalkanoates (PHA), proteins including surfactants and enzymes, small molecular compounds including amino acids and derivates, as well as organic acids. This paper reviews all the progress reported in the last 10 years regarding this robust microbial cell factory.
Key points
• Halomonas spp. are robust chassis for low-cost production of chemicals
• Genomic information of some Halomonas spp. has been revealed
• Molecular tools and approaches for Halomonas spp. have been developed
• Halomonas spp. are becoming more and more important for biotechnology


Similar content being viewed by others
References
Afendra AS, Vargas C, Nieto JJ, Drainas C (2004) Gene transfer and expression of recombinant proteins in moderately halophilic bacteria. In: Balbás P, Lorence A (eds) Recombinant gene expression, Methods in Molecular Biology, vol 267, Humana Press, pp 209–223
Argandoña MVC, Reina-Bueno M, Rodríguez-Moya J, Salvador M and Nieto J (2012) An extended suite of genetic tools for use in bacteria of the Halomonadaceae: an overview. Methods Mol Biol 824:167–201
Becker J, Zelder O, Hafner S, Schroder H, Wittmann C (2011) From zero to hero–design-based systems metabolic engineering of Corynebacterium glutamicum for L-lysine production. Metab Eng 13:159–168
Cai L, Tan D, Aibaidula G, Dong XR, Chen JC, Tian WD and Chen GQ (2011) Comparative genomics study of polyhydroxyalkanoates (PHA) and ectoine relevant genes from Halomonas sp. TD01 revealed extensive horizontal gene transfer events and co-evolutionary relationships. Microb Cell Fact 10: 88
Camacho DM, Collins KM, Powers RK, Costello JC, Collins JJ (2018) Next-generation machine learning for biological networks. Cell 173:1581–1592
Chen Y, Chen XY, Du HT, Zhang X, Ma YM, Chen JC, Ye JW, Jiang XR, Chen GQ (2019) Chromosome engineering of the TCA cycle in Halomonas bluephagenesis for production of copolymers of 3-hydroxybutyrate and 3-hydroxyvalerate (PHBV). Metab Eng 54:69–82
Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
DasSarma SL, Capes MD, DasSarma P, DasSarma S (2010) HaloWeb: the haloarchaeal genomes database. Saline Syst 6:12
Don TM, Chen CW, Chan TH (2006) Preparation and characterization of poly(hydroxyalkanoate) from the fermentation of Haloferax mediterranei. J Biomater Sci Polym Ed 17:1425–1438
Dong X, Quinn PJ, Wang X (2011) Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for the production of L-threonine. Biotechnol Adv 29:11–23
Douka ECA, Koukkou AI, Afendra AS, Drainas C (2001) Use of a green fluorescent protein gene as a reporter in Zymomonas mobilis and Halomonas elongata. FEMS Microbiol Lett 201:221–227
Du H, Zhao Y, Wu F, Ouyang P, Chen J, Jiang X, Ye J, Chen GQ (2020) Engineering Halomonas bluephagenesis for L-Threonine production. Metab Eng 60:119–127
Fu XZ, Tan D, Aibaidula G, Wu Q, Chen JC, Chen GQ (2014) Development of Halomonas TD01 as a host for open production of chemicals. Metab Eng 23:78–91
Galinski EA, PFEIFFER HP, Trüper HG (1985) 1,4,5,6-Tetrahydro-2-methyl-4-pyrimidinecarboxylic acid. A novel cyclic amino acid from halophilic phototrophic bacteria of the genus Ectothiorhodospira. Eur J Biochem 149: 135–139
Glaser GSP, Cashel M (1983) Functional interrelationship between two tandem E. coli ribosomal RNA promoters. Nature 302:74–76
Goh S (2016) Phage transduction. In: Roberts A, Mullany P (eds) Clostridium difficile, Methods in Molecular Biology, vol 1476. Humana Press, pp 177–185
Gonzalez O, Gronau S, Falb M, Pfeiffer F, Mendoza E, Zimmer R, Oesterhelt D (2008) Reconstruction, modeling & analysis of Halobacterium salinarum R-1 metabolism. Mol BioSyst 4:148–159
Grammann K, Volke A, Kunte HJ (2002) New type of osmoregulated solute transporter identified in halophilic members of the bacteria domain: TRAP transporter TeaABC mediates uptake of ectoine and hydroxyectoine in Halomonas elongata DSM 2581(T). J Bacteriol 184:3078–3085
Gupta A, Reizman IM, Reisch CR, Prather KL (2017) Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat Biotechnol 35:273–279
Han JLQ, Zhou L, Zhou J, Xiang H (2007) Molecular characterization of the phaECHm genes, required for biosynthesis of poly(3-hydroxybutyrate) in the extremely halophilic archaeon Haloarcula marismortui. Appl Environ Microbiol 73:6058–6606
Harris JR, Lundgren BR, Grzeskowiak BR, Mizuno K, Nomura CT (2016) A rapid and efficient electroporation method for transformation of Halomonas sp. O-1. J Microbiol Methods 129:127–132
Hayes F (2003) Transposon-based strategies for microbial functional genomics and proteomics. Annu Rev Genet 37:3–29
Jiang XR, Yao ZH, Chen GQ (2017) Controlling cell volume for efficient PHB production by Halomonas. Metab Eng 44:30–37
Jiang XR, Yan X, Yu LP, Liu XY, Chen GQ (2021) Hyperproduction of 3-hydroxypropionate by Halomonas bluephagenesis. Nat Commun 12:1513
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Jing Y, Bian Y, Hu Z, Wang L, Xie X-QS (2018) Deep learning for drug design: an artificial intelligence paradigm for drug discovery in the big data era. AAPS J 20:58
Joshi AA, Kanekar PP, Kelkar AS, Sarnaik SS, Shouche Y, Wani A (2007) Moderately halophilic, alkalitolerant Halomonas campisalis MCM B-365 from Lonar Lake, India. J Basic Microb 47:213–221
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589
Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176
Kraegeloh A, Amendt B, Kunte HJ (2005) Potassium transport in a halophilic member of the Bacteria domain: identification and characterization of the K+ uptake systems TrkH and TrkI from Halomonas elongata DSM 2581(T). J Bacteriol 187:1036–1043
Kunte HJ, Galinski EA (1995) Transposon mutagenesis in halophilic eubacteria: conjugal transfer and insertion of transposon Tn5 and Tn1732 in Halomonas elongata. FEMS Microbiol Lett 128:293–299
Lach J, Jęcz P, Strapagiel D, Matera-Witkiewicz A, Stączek P (2021) The methods of digging for “Gold” within the salt: characterization of halophilic prokaryotes and identification of their valuable biological products using sequencing and genome mining tools. Genes 12:1756
Lan LH, Zhao H, Chen JC, Chen GQ (2016) Engineering Halomonas spp. as a low-cost production host for production of bio-surfactant protein PhaP. Biotechnol J 11:1595–1604
Li T, Guo YY, Qiao GQ, Chen GQ (2016a) Microbial synthesis of 5-aminolevulinic acid and its coproduction with polyhydroxybutyrate. ACS Synth Biol 5:1264–1274
Li T, Ji W, Wang Q, Zhang H, Chen GQ, Lou CB, Ouyang Q (2016b) Engineering of core promoter regions enables the construction of constitutive and inducible promoters in Halomonas sp. Biotechnol J 11:219–227
Li T, Ye JW, Shen R, Zong YQ, Zhao X, Lou C, Chen GQ (2016c) Semirational approach for ultrahigh poly(3-hydroxybutyrate) accumulation in Escherichia coli by combining one-step library construction and high-throughput screening. ACS Synth Biol 5:1308–1317
Lin Y, Guan Y, Dong X, Ma Y, Wang X, Leng Y, Wu F, Ye JW, Chen GQ (2021) Engineering Halomonas bluephagenesis as a chassis for bioproduction from starch. Metab Eng 64:134–145
Ling C, Qiao GQ, Shuai B-W, Olavarria K, Yin J, Xiang RJ, Song KN, Shen YH, Guo Y, Chen GQ (2018) Engineering NADH/NAD+ ratio in Halomonas bluephagenesis for enhanced production of polyhydroxyalkanoates (PHA). Metab Eng 49:275–286
Loukas A, Kappas I, Abatzopoulos TJ (2018) HaloDom: a new database of halophiles across all life domains. J Biol Res Thessalon 25:2
Ma HK, Liu MM, Li SY, Wu Q, Chen JC, Chen GQ (2013) Application of polyhydroxyalkanoate (PHA) synthesis regulatory protein PhaR as a bio-surfactant and bactericidal agent. J Biotechnol 166:34–41
Ma H, Zhao Y, Huang W, Zhang L, Wu F, Ye J, Chen GQ (2020) Rational flux-tuning of Halomonas bluephagenesis for co-production of bioplastic PHB and ectoine. Nat Commun 11:3313
Ma Y, Zheng X, Lin Y, Zhang L, Yuan Y, Wang H, Winterburn J, Wu F, Wu Q, Ye JW, Chen GQ (2022) Engineering an oleic acid-induced system for Halomonas, E. Coli and Pseudomonas. Metab Eng 72:325–336
Mali P, Yang LH, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826
Oren A (2008) Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Syst 4:2
Ouyang PF, Wang H, Hajnal I, Wu Q, Guo YY, Chen GQ (2018) Increasing oxygen availability for improving poly(3-hydroxybutyrate) production by Halomonas. Metab Eng 45:20–31
Oyewusi, HA, Akinyede KA, Abdul Wahab R and Huyop F (2021) In silico analysis of a putative dehalogenase from the genome of halophilic bacterium Halomonas smyrnensis AAD6T. J Biomol Struct Dyn 1–17
Qin Q, Ling C, Zhao Y, Yang T, Yin J, Guo Y, Chen GQ (2018) CRISPR/Cas9 editing genome of extremophile Halomonas spp. Metab Eng 47:219–229
Rensing C (2005) Adaption to life at high salt concentrations in Archaea. Bacteria and Eukarya Saline Syst 1:6
Rivera-Terceros P, Tito-Claros E, Torrico S, Carballo S, Van-Thuoc D, Quillaguamán J (2015) Production of poly(3-hydroxybutyrate) by Halomonas boliviensis in an air-lift reactor. J Biol Res-Thessalon 22:8
Schafer R, Zillig W (1973) The effects of ionic strength on termination of transcription of DNAs from bacteriophages T4, T5 and T7 by DNA-dependent RNA polymerase from Escherichia coli, and the nature of termination by factor ρ. Eur J Biochem 33:215–226
Seaman PF, Day MJ (2007) Isolation and characterization of a bacteriophage with an unusually large genome from the Great Salt Plains National Wildlife Refuge, Oklahoma, USA. FEMS Microbiol Ecol 60:1–13
Sharma N, Farooqi MS, Chaturvedi KK, Lal SB, Grover M, Rai A, Pandey P (2014) The halophile protein database. Database (Oxford) 2014: bau114-bau114
Shen R, Yin J, Ye JW, Xiang RJ, Ning ZY, Huang WZ, Chen GQ (2018) Promoter engineering for enhanced P(3HB-co-4HB) production by Halomonas bluephagenesis. ACS Synth Biol 7:1897–1906
Silva-Rocha R, Martinez-Garcia E, Calles B, Chavarria M, Arce-Rodriguez A, de Las Heras A, Paez-Espino AD, Durante-Rodriguez G, Kim J, Nikel PI, Platero R, Lorenzo Vd (2013) The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res 41:D666-675
Sogutcu E, Emrence Z, Arikan M, Cakiris A, Abaci N, Öner Ebru T, Üstek D, Arga Kazim Y (2012) Draft genome sequence of Halomonas smyrnensis AAD6T. J Bacteriol 194:5690–5691
Tan D, Xue YS, Aibaidula G, Chen GQ (2011) Unsterile and continuous production of polyhydroxybutyrate by Halomonas TD01. Bioresource Technol 102:8130–8136
Tan D, Wu Q, Chen JC, Chen GQ (2014) Engineering Halomonas TD01 for the low-cost production of polyhydroxyalkanoates. Metab Eng 26:34–47
Tao W, Lv L, Chen GQ (2017) Engineering Halomonas species TD01 for enhanced polyhydroxyalkanoates synthesis via CRISPRi. Microb Cell Fact 16:48
Thomas T, Elain A, Bazire A and Bruzaud S (2019) Complete genome sequence of the halophilic PHA-producing bacterium Halomonas sp. SF2003: insights into its biotechnological potential. World J Microbiol Biotechnol 35: 50
Tokunaga M, Arakawa T, Tokunaga H (2010) Recombinant expression in moderate halophiles. Curr Pharm Biotechnol 11:259–266
Ukani H, Purohit MK, Georrge JJ, Paul S, Singh SP (2011) HaloBase: development of database system for halophilic bacteria and archaea with respect to proteomics, genomics & other molecular traits. J Sci Ind Res India 70:976–981
Wang Y, Wang H, Wei L, Li S, Liu L, Wang X (2020) Synthetic promoter design in Escherichia coli based on a deep generative network. Nucleic Acids Res 48:6403–6412
Wang Z, Qin Q, Zheng Y, Li F, Zhao Y, Chen GQ (2021) Engineering the permeability of Halomonas bluephagenesis enhanced its chassis properties. Metab Eng 67:53–66
Wang L-J, Jiang X-R, Hou J, Wang C-H, Chen G-Q (2022) Engineering Halomonas bluephagenesis via small regulatory RNAs. Metab Eng 73:58–69
Xu T, Chen JY, Mitra R, Lin L, Xie ZW, Chen GQ, Xiang H, Han J (2022) Deficiency of exopolysaccharides and O-antigen makes Halomonas bluephagenesis self-flocculating and amenable to electrotransformation. Communication Biol 5:623
Ye J-W, Chen G-Q (2021) Halomonas as a chassis. Essays Biochem 65:393–403
Ye JW, Hu DK, Yin J, Huang WZ, Xiang RJ, Zhang LZ, Wang X, Han JN, Chen GQ (2020) Stimulus response-based fine-tuning of polyhydroxyalkanoate pathway in Halomonas. Metab Eng 57:85–95
Yin J, Fu XZ, Wu Q, Chen JC, Chen GQ (2014) Development of an enhanced chromosomal expression system based on porin synthesis operon in Halomonas TD01. Appl Microbiol Biotechnol 98:8987–8997
Yu LP, Wu FQ, Chen GQ (2019) Next-generation industrial biotechnology-transforming the current industrial biotechnology into competitive processes. Biotechnol J 14:1800437
Yu LP, Yan X, Zhang X, Chen XB, Wu Q, Jiang XR, Chen GQ (2020) Biosynthesis of functional polyhydroxyalkanoates by engineered Halomonas bluephagenesis. Metab Eng 59:119–130
Zhang J, Zhang X, Mao Y, Jin B, Guo Y, Wang Z, Chen T (2020a) Substrate profiling and tolerance testing of Halomonas TD01 suggest its potential application in sustainable manufacturing of chemicals. J Biotechnol 316:1–5
Zhang X, Lin Y, Wu Q, Wang Y, Chen GQ (2020b) Synthetic biology and genome-editing tools for improving PHA metabolic engineering. Trends Biotechnol 38:689–700
Zhang J, Jin B, Hong K, Lv Y, Wang Z, Chen T (2021) Cell catalysis of citrate to itaconate by engineered Halomonas bluephagenesis. ACS Synth Biol 10:3017–3027
Zhang L, Ye J-W, Zhang X, Huang W, Zhang Z, Lin Y, Zhang G, Wu F, Wang Z, Wu Q, Chen G-Q (2022) Effective production of Poly(3-hydroxybutyrate-co-4-hydroxybutyrate) by engineered Halomonas bluephagenesis grown on glucose and 1,4-Butanediol. Bioresource Technol 355:127270
Zhao H, Zhang HQM, Li T, Lou CB, Ouyang Q, Chen GQ (2017) Novel T7-like expression systems used for Halomonas. Metab Eng 39:128–140
Zhao C, Zheng T, Feng Y, Wang X, Zhang L, Hu Q, Chen J, Wu F, Chen GQ (2022) Engineered Halomonas spp for production of L-lysine and cadaverine. Bioresource Technol 349:126865
Zou J, Huss M, Abid A, Mohammadi P, Torkamani A, Telenti A (2019) A primer on deep learning in genomics. Nat Genet 51:12–18
Acknowledgements
We are grateful for the kind donation of pSEVA plasmids from Professor Victor de Lorenzo of Ctr Nacl Biotecnol CSIC/Spain.
Funding
This work was financially supported by grants from the Ministry of Science and Technology of China (Grant No. 2018YFA0900200), National Natural Science Foundation of China (Grant No. 21761132013; No. 31870859; No. 92068117), Tsinghua University-INDITEX Sustainable Development Fund (Grant No. TISD201907), and Center of Life Sciences of Tsinghua-Peking University. This project was also funded by the National Natural Science Foundation of China (Grant No. 31961133017; No. 31961133018; No. 31961133019). These grants are part of MIX-UP, a joint NSFC and EU H2020 collaboration. In Europe, MIX-UP has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement no. 870294.
Author information
Authors and Affiliations
Contributions
GQC designed, conceptualized, edited, and supervised the entire work. XZ, WH, JH, and JZ prepared the primary draft. XZ, ZX, CZ, and TC revised it and designed the figures. XL, TX, and RM prepared the tables. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, GQ., Zhang, X., Liu, X. et al. Halomonas spp., as chassis for low-cost production of chemicals. Appl Microbiol Biotechnol 106, 6977–6992 (2022). https://doi.org/10.1007/s00253-022-12215-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-12215-3