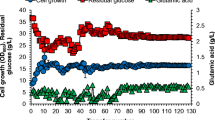
Abstract
Yarrowia lipolytica strain is a promising cell factory for the conversion of lignocellulose to biofuels and bioproducts. Despite the inherent robustness of this strain, further improvements to lignocellulose-derived inhibitors toxicity tolerance of Y. lipolytica are also required to achieve industrial application. Here, adaptive laboratory evolution was employed with increasing concentrations of ferulic acid. The adaptive laboratory evolution experiments led to evolve Y. lipolytica strain yl-XYL + *FA*4 with increased tolerance to ferulic acid as compared to the parental strain. Specifically, the evolved strain could tolerate 1.5 g/L ferulic acid, whereas 0.5 g/L ferulic acid could cause about 90% lethality of the parental strain. Transcriptome analysis of the evolved strain revealed several targets underlying toxicity tolerance enhancements. YALI0_E25201g, YALI0_F05984g, YALI0_B18854g, and YALI0_F16731g were among the highest upregulated genes, and the beneficial contributions of these genes were verified via reverse engineering. Recombinant strains with overexpressing each of these four genes obtained enhanced tolerance to ferulic acid as compared to the control strain. Fortunately, recombinant strains with overexpression of YALI0_E25201g, YALI0_B18854g, and YALI0_F16731g individually also obtained enhanced tolerance to vanillic acid. Overall, this work demonstrated a whole strain improvement cycle by “non-rational” metabolic engineering and presented new targets to modify Y. lipolytica for microbial lignocellulose valorization.
Key points
• Adaptive evolution improved the ferulic acid tolerance of Yarrowia lipolytica
• Transcriptome sequence was applied to analyze the ferulic acid tolerate strain
• Three genes were demonstrated for both ferulic acid and vanillic acid tolerance







Similar content being viewed by others
Data availability
The datasets of RNA sequencing raw data generated in this study have been submitted in Genbank with an accession number of PRJNA 675399 (https://dataview.ncbi.nlm.nih.gov/object/PRJNA675399).
References
Allen SA, Clark W, Mccaffery JM, Zhen C, Gorsich SW (2010) Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnol Biofuels 3:1–10. https://doi.org/10.1186/1754-6834-3-2
Almario M, Reyes L, Kao K (2013) Evolutionary engineering of Saccharomyces cerevisiae for enhanced tolerance to hydrolysates of lignocellulosic biomass. Biotechnol Bioeng 110. 2616-2623. doi:https://doi.org/10.1002/bit.24938
Almeida JR, Modig T, Petersson A, Hähn-Hägerdal B, Lidén G, Gorwa-Grauslund MF (2010) Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J Chem Technol Biotechnol 82:340–349. https://doi.org/10.1002/jctb.1676
Ask M, Mapelli V, Hck H, Olsson L, Bettiga M (2013) Engineering glutathione biosynthesis of Saccharomyces cerevisiae increases robustness to inhibitors in pretreated lignocellulosic materials. Microb Cell Factories 12:87. https://doi.org/10.1186/1475-2859-12-87
Brandt BA, Jansen T, Görgens JF, van Zyl WH (2019) Overcoming lignocellulose-derived microbial inhibitors: advancing the Saccharomyces cerevisiae resistance toolbox. Biofuels Bioprod Biorefin 13:1520–1536. https://doi.org/10.1002/bbb.2042
Chen H, Li J, Wan C, Fang Q, Zhao X (2019) Improvement of inhibitor tolerance in Saccharomyces cerevisiae by overexpression of the quinone oxidoreductase family gene YCR102C. FEMS Yeast Res 19:55. https://doi.org/10.1093/femsyr/foz055
Chen X, Zhai R, Shi K, Yuan Y, Dale BE, Gao Z, Jin M (2018) Mixing alkali pretreated and acid pretreated biomass for cellulosic ethanol production featuring reduced chemical use and decreased inhibitory effect. Ind Crop Prod 124:719–725. https://doi.org/10.1016/j.indcrop.2018.08.056
Cho DH, Lee YJ, Um Y, Sang BI, Kim YH (2009) Detoxification of model phenolic compounds in lignocellulosic hydrolysates with peroxidase for butanol production from Clostridium beijerinckii. Appl Microbiol Biotechnol 83:1035–1043. https://doi.org/10.1007/s00253-009-1925-8
Daskalaki A, Perdikouli N, Aggeli D, Aggelis G (2019) Laboratory evolution strategies for improving lipid accumulation in Yarrowia lipolytica. Appl Microbiol Biotechnol 103:8585–8596. https://doi.org/10.1007/s00253-019-10088-7
Dragosits M, Mattanovich D (2013) Adaptive laboratory evolution–principles and applications for biotechnology. Microb Cell Factories 12:64. https://doi.org/10.1186/1475-2859-12-64
Gao S, Tong Y, Wen Z, Zhu L, Ge M, Chen D, Jiang Y, Yang S (2016) Multiplex gene editing of the Yarrowia lipolytica genome using the CRISPR-Cas9 system. J Ind Microbiol Biotechnol 43:1085–1093. https://doi.org/10.1007/s10295-016-1789-8
Guo X, Cavka A, Jönsson LJ, Hong F (2013) Comparison of methods for detoxification of spruce hydrolysate for bacterial cellulose production. Microb Cell Factories 12:93. https://doi.org/10.1186/1475-2859-12-93
Heipieper HJ, Weber FJ, Sikkema J, Keweloh H, de Bont JA (1994) Mechanisms of resistance of whole cells to toxic organic solvents. Trends Biotechnol 12:409–415. https://doi.org/10.1016/0167-7799(94)90029-9
Hellgren J (2017) Adaptive evolution of Yarrowia lipolytica for osmotic and saline tolerance. Dissertation, Chalmers University Of Technology
Hou J, Ding C, Qiu Z, Zhang Q, Xiang WN (2017) Inhibition efficiency evaluation of lignocellulose-derived compounds for bioethanol production. J Clean Prod 165:1107–1114. https://doi.org/10.1016/j.jclepro.2017.07.204
Imai T, Ohno T (1995) The relationship between viability and intracellular pH in the yeast Saccharomyces cerevisiae. Appl Environ Microbiol 61:3604–3608. https://doi.org/10.1002/bit.260480111
Jacobsen IH, Ledesma-Amaro R, Martinez JL (2020) Recombinant β-Carotene production by Yarrowia lipolytica – assessing the potential of micro-scale fermentation analysis in cell factory design and bioreaction optimization. Front Bioeng Biotech 8. 29. doi:https://doi.org/10.3389/fbioe.2020.00029
Jin M, Slininger PJ, Dien BS, Waghmode S, Moser BR, Orjuela A, Sousa LC, Balan V (2015) Microbial lipid-based lignocellulosic biorefinery: feasibility and challenges. Trends Biotechnol 33:43–54. https://doi.org/10.1016/j.tibtech.2014.11.005
Klinke HB, Thomsen AB, Ahring BK (2004) Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl Microbiol Biotechnol 66:10–26. https://doi.org/10.1007/s00253-004-1642-2
Kurosawa K, Laser J, Sinskey AJ (2015) Tolerance and adaptive evolution of triacylglycerol-producing Rhodococcus opacus to lignocellulose-derived inhibitors. Biotechnol Biofuels 8:76. https://doi.org/10.1186/s13068-015-0258-3
Larroude M, Celinska E, Back A, Thomas S, Nicaud JM, Ledesma-Amaro R (2017) A synthetic biology approach to transform Yarrowia lipolytica into a competitive biotechnological producer of β-carotene. Biotechnol Bioeng 115(2):464–472. https://doi.org/10.1002/bit.26473
Larsson S, Quintana-Sáinz A, Reimann A, Nilvebrant NO, Jönsson LJ (2000) Influence of lignocellulose-derived aromatic compounds on oxygen-limited growth and ethanolic fermentation by Saccharomyces cerevisiae. Appl Biochem Biotechnol 84-86:617–632. https://doi.org/10.1385/abab:84-86:1-9:617
Ledesma-Amaro R, Lazar Z, Rakicka M, Guo Z, Fouchard F, Coq CL, Nicaud JM (2016) Metabolic engineering of Yarrowia lipolytica to produce chemicals and fuels from xylose. Metab Eng 38:115–124. https://doi.org/10.1016/j.ymben.2016.07.001
Liu Y, Jiang X, Cui Z, Wang Z, Hou J (2019) Engineering the oleaginous yeast Yarrowia lipolytica for production of α-farnesene. Biotechnol Biofuels 12:296. https://doi.org/10.1186/s13068-019-1636-z
López MJ, Nichols NN, Dien BS, Moreno J, Bothast RJ (2004) Isolation of microorganisms for biological detoxification of lignocellulosic hydrolysates. Appl Microbiol Biotechnol 64:125–131. https://doi.org/10.1007/s00253-003-1401-9
Mansfield SD, Kim H, Lu F, Ralph J (2012) Whole plant cell wall characterization using solution-state 2D NMR. Nat Protoc 7:1579–1589. https://doi.org/10.1038/nprot.2012.064
Modig T, Lidén G, Taherzadeh MJ (2002) Inhibition effects of furfural on alcohol dehydrogenase, aldehyde dehydrogenase and pyruvate dehydrogenase. Biochem J 363:769–776. https://doi.org/10.1042/0264-6021:3630769
Pampulha ME, Loureiro-Dias MC (1990) Activity of glycolytic enzymes of Saccharomyces cerevisiae in the presence of acetic acid. Appl Microbiol Biotechnol 34:375–380. https://doi.org/10.1007/BF00170063
Qi K, Xia X, Zhong J (2015) Enhanced anti-oxidative activity and lignocellulosic ethanol production by biotin addition to medium in Pichia guilliermondii fermentation. Bioresour Technol 189:36–43. https://doi.org/10.1016/j.biortech.2015.02.089
Qiao K, Wasylenko TM, Zhou K, Xu P, Stephanopoulos G (2017) Lipid production in Yarrowia lipolytica is maximized by engineering cytosolic redox metabolism. Nat Biotechnol 35:173–177. https://doi.org/10.1038/nbt.3763
Qiu Z, Deng Z, Tan H, Zhou S, Cao L (2015) Engineering the robustness of Saccharomyces cerevisiae by introducing bifunctional glutathione synthase gene. J Ind Microbiol Biotechnol 42:537–542. https://doi.org/10.1007/s10295-014-1573-6
Ruan Z, Zanotti M, Wang X, Ducey C, Liu Y (2012) Evaluation of lipid accumulation from lignocellulosic sugars by Mortierella isabellina for biodiesel production. Bioresour Technol 110:198–205. https://doi.org/10.1016/j.biortech.2012.01.053
Ruan Z, Zanotti M, Zhong Y, Liao W, Ducey C, Liu Y (2013) Co-hydrolysis of lignocellulosic biomass for microbial lipid accumulation. Biotechnol Bioeng 110:1039–1049. https://doi.org/10.1002/bit.24773
Sandberg TE, Salazar MJ, Weng LL, Palsson BO, Feist AM (2019) The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab Eng 56:1–16. https://doi.org/10.1016/j.ymben.2019.08.004
Shen H, Zhang X, Gong Z, Wang Y, Yu X, Yang X, Zhao Z (2017) Compositional profiles of Rhodosporidium toruloides cells under nutrient limitation. Appl Microbiol Biotechnol 101:3801–3809. https://doi.org/10.1007/s00253-017-8157-0
Shishir PSC, Gregg TB, Michael EH, Bruce ED (2011) Deconstruction of lignocellulosic biomass to fuels and Cchemicals. Annu Rev Chem Biomol Eng 2:121–145. https://doi.org/10.1146/annurev-chembioeng-061010-114205
Thompson OA, Hawkins GM, Gorsich SW, Doran-Peterson J (2016) Phenotypic characterization and comparative transcriptomics of evolved Saccharomyces cerevisiae strains with improved tolerance to lignocellulosic derived inhibitors. Biotechnol Biofuels 9:200. https://doi.org/10.1186/s13068-016-0614-y
Wang X, Khushk I, Xiao Y, Gao Q, Bao J (2018) Tolerance improvement of Corynebacterium glutamicum on lignocellulose derived inhibitors by adaptive evolution. Appl Microbiol Biotechnol 102:377–388. https://doi.org/10.1007/s00253-017-8627-4
Wei L, Cao L, Miao Y, Wu S, Xu S, Wang R, Du J, Liang A, Fu Y (2017) Transcriptome analysis of Spodoptera frugiperda 9 (Sf9) cells infected with baculovirus, AcMNPV or AcMNPV-BmK IT. Biotechnol Lett 39:1129–1139. https://doi.org/10.1007/s10529-017-2356-8
Wen Z, Wu M, Lin Y, Yang L, Lin J, Cen P (2014) Artificial symbiosis for acetone-butanol-ethanol (ABE) fermentation from alkali extracted deshelled corn cobs by co-culture of Clostridium beijerinckii and Clostridium cellulovorans. Microb Cell Factories 13:1–11. https://doi.org/10.1186/s12934-014-0092-5
Wu D, Wang D, Hong J (2020) Effect of a novel alpha/beta hydrolase domain protein on tolerance of K. marxianus to lignocellulosic biomass derived inhibitors. Front Bioeng Biotechnol 8:844. https://doi.org/10.3389/fbioe.2020.00844
Wu Y, Wang T, Wang K, Liang Q, Bai Z, Liu Q, Pan Y, Jiang B, Zhang L (2016) Comparative analysis of the chrysanthemum leaf transcript profiling in response to salt stress. PLoS One 11:e0159721. https://doi.org/10.1371/journal.pone.0159721
Xu J, Liu N, Qiao K, Vogg S, Stephanopoulos G (2017) Application of metabolic controls for the maximization of lipid production in semicontinuous fermentation. Proc Natl Acad Sci U S A 114:E5308–E5316. https://doi.org/10.1073/pnas.1703321114
Xu Z, Lei P, Zhai R, Wen Z, Jin M (2019) Recent advances in lignin valorization with bacterial cultures: microorganisms, metabolic pathways, and bio-products. Biotechnol Biofuels 12:32. https://doi.org/10.1186/s13068-019-1376-0
Yang X, Nambou K, Wei L, Hua Q (2016) Heterologous production of α-farnesene in metabolically engineered strains of Yarrowia lipolytica. Bioresour Technol 216:1040–1048. https://doi.org/10.1016/j.biortech.2016.06.028
Yook SD, Kim J, Gong G, Ko JK, Um Y, Han SO, Lee SM (2020) High-yield lipid production from lignocellulosic biomass using engineered xylose-utilizing Yarrowia lipolytica. GCB Bioenergy 12:670–679. https://doi.org/10.1111/gcbb.12699
Yu X, Zheng Y, Dorgan KM, Chen S (2011) Oil production by oleaginous yeasts using the hydrolysate from pretreatment of wheat straw with dilute sulfuric acid. Bioresour Technol 102:6134–6140. https://doi.org/10.1016/j.biortech.2011.02.081
Zhou L, Wen Z, Wang Z, Zhang Y, Ledesma-Amaro R, Jin M (2020) Evolutionary Engineering Improved d-Glucose/Xylose Cofermentation of Yarrowia lipolytica. Ind Eng Chem Res 59:17113–17123. https://doi.org/10.1021/acs.iecr.0c00896
Acknowledgements
We would like to thank Dr. Rodrigo Ledesma-Amaro for providing Y. lipolytica yl-XYL+.
Funding
This work was supported by “Natural Science Foundation of Jiangsu Province,” Grant No. BK20170037 and BK20170829; “National Key R&D Program of China,” Grant No. 2016YFE0105400; “National Natural Science Foundation of China,” Grant Nos. 21606132 and 21706133.
Author information
Authors and Affiliations
Contributions
JM and WZ conceived and designed research. WZ, ZL, WZ, ZY, LM, and ZH performed the experiments; WZ, ZL, XZ, and WZ analyzed the data; JM, XZ, and WZ coordinated and supervised this study; WZ drafted the manuscript. ZL, XZ, and JM revised the manuscript.
Corresponding authors
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals that were performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, Z., Zhou, L., Lu, M. et al. Adaptive laboratory evolution of Yarrowia lipolytica improves ferulic acid tolerance. Appl Microbiol Biotechnol 105, 1745–1758 (2021). https://doi.org/10.1007/s00253-021-11130-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11130-3