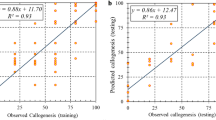
Abstract
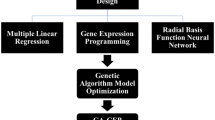
Data-driven models in a combination of optimization algorithms could be beneficial methods for predicting and optimizing in vitro culture processes. This study was aimed at modeling and optimizing a new embryogenesis medium for chrysanthemum. Three individual data-driven models, including multi-layer perceptron (MLP), adaptive neuro-fuzzy inference system (ANFIS), and support vector regression (SVR), were developed for callogenesis rate (CR), embryogenesis rate (ER), and somatic embryo number (SEN). Consequently, the best obtained results were used in the fusion process by a bagging method. For medium reformulation, effects of eight ionic macronutrients on CR, ER, and SEN and effects of four vitamins on SEN were evaluated using data fusion (DF)–non-dominated sorting genetic algorithm-II (NSGA-II) and DF-genetic algorithm (GA), respectively. Results showed that DF models with the highest R2 had superb performance in comparison with all other individual models. According to DF-NSGAII, the highest ER and SEN can be obtained from the medium containing 14.27 mM NH4+, 38.92 mM NO3−, 22.79 mM K+, 5.08 mM Cl−, 3.34 mM Ca2+, 1.67 mM Mg2+, 2.17 mM SO42−, and 1.44 mM H2PO4−. Based on the DF-GA model, the maximum SEN can be obtained from a medium containing 0.61 μM thiamine, 5.93 μM nicotinic acid, 0.25 μM biotin, and 0.26 μM riboflavin. The efficiency of the established-optimized medium was experimentally compared to Murashige and Skoog medium (MS) for embryogenesis of five chrysanthemum cultivars, and results indicated the efficiency of optimized medium over MS medium.
Key points
• MLP, SVR, and ANFIS were fused by a bagging method to develop a data fusion model.
• NSGA-II and GA were linked to the data fusion model for establishing and optimizing a new embryogenesis medium.
• The new culture medium (HNT) had better efficiency than MS medium.

Similar content being viewed by others
Data availability
All processed data are available without restriction upon inquiry.
References
Aiello G, Giovino I, Vallone M, Catania P, Argento A (2018) A decision support system based on multisensor data fusion for sustainable greenhouse management. J Clean Prod 172:4057–4065. https://doi.org/10.1016/j.jclepro.2017.02.197
Akin M, Eyduran SP, Eyduran E, Reed BM (2020) Analysis of macro nutrient related growth responses using multivariate adaptive regression splines. Plant Cell Tissue Organ Cult 140:661–670. https://doi.org/10.1007/s11240-019-01763-8
Al-Khayri JM (2001) Optimization of biotin and thiamine requirements for somatic embryogenesis of date palm (Phoenix dactylifera L.). In Vitro Cell Dev Biol Plant 37(4):453–456. https://doi.org/10.1007/s11627-001-0079-x
Alanagh EN, G-a G, Haddad R, Maleki S, Landín M, Gallego PP (2014) Design of tissue culture media for efficient Prunus rootstock micropropagation using artificial intelligence models. Plant Cell Tissue Organ Cult 117(3):349–359. https://doi.org/10.1007/s11240-014-0444-1
Alizadeh MR, Nikoo MR (2018) A fusion-based methodology for meteorological drought estimation using remote sensing data. Remote Sens Environ 211:229–247. https://doi.org/10.1016/j.rse.2018.04.001
Arab MM, Yadollahi A, Ahmadi H, Eftekhari M, Maleki M (2017) Mathematical modeling and optimizing of in vitro hormonal combination for G× N15 vegetative rootstock proliferation using artificial neural network-genetic algorithm (ANN-GA). Front Plant Sci 8:1853. https://doi.org/10.3389/fpls.2017.01853
Araghinejad S, Fayaz N, Hosseini-Moghari S-M (2018) Development of a hybrid data driven model for hydrological estimation. Water Resour Manag 32(11):3737–3750. https://doi.org/10.1007/s11269-018-2016-3
Araghinejad S, Hosseini-Moghari S-M, Eslamian S (2017) Application of data-driven models in drought forecasting. In: Handbook of Drought and Water Scarcity. CRC Press, pp. 423–440
Arora DK, Sun SS, Ramawat KG, Merillon J-M (1999) Factors affecting somation embryogenesis in long term callus cultures of ‘safed musli’ (Chlorophytum borivilianum), an endangered wonder herb. Indian J Exp Biol 31:75–82
Arruda S, Souza G, Almeida M, Gonçalves A (2000) Anatomical and biochemical characterization of the calcium effect on Eucalyptus urophylla callus morphogenesis in vitro. Plant Cell Tissue Organ Cult 63(2):142–154. https://doi.org/10.1023/A:1006482702094
Asano Y, Katsumoto H, Inokuma C, Kaneko S, Ito Y, Fujiie A (1996) Cytokinin and thiamine requirements and stimulative effects of riboflavin and α-ketoglutaric acid on embryogenic callus induction from the seeds of Zoysia japonica Steud. J Plant Physiol 149(3–4):413–417. https://doi.org/10.1016/S0176-1617(96)80142-8
Asensi-Fabado MA, Munné-Bosch S (2010) Vitamins in plants: occurrence, biosynthesis and antioxidant function. Trends Plant Sci 15(10):582–592. https://doi.org/10.1016/j.tplants.2010.07.003
Ashihara H, Ludwig IA, Katahira R, Yokota T, Fujimura T, Crozier A (2015) Trigonelline and related nicotinic acid metabolites: occurrence, biosynthesis, taxonomic considerations, and their roles in planta and in human health. Phytochem Rev 14(5):765–798. https://doi.org/10.1007/s11101-014-9375-z
Barcelo P, Lazzeri PA, Martin A, Lörz H (1992) Competence of cereal leaf cells. II. Influence of auxin, ammonium and explant age on regeneration. J Plant Physiol 139(4):448–454. https://doi.org/10.1016/S0176-1617(11)80493-1
Barone JO (2019) Use of multiple regression analysis and artificial neural networks to model the effect of nitrogen in the organogenesis of Pinus taeda L. Plant Cell Tissue Organ Cult 137(3):455–464. https://doi.org/10.1007/s11240-019-01581-y
Barwale U, Kerns H, Widholm J (1986) Plant regeneration from callus cultures of several soybean genotypes via embryogenesis and organogenesis. Planta 167(4):473–481. https://doi.org/10.1007/BF00391223
Bonner J (1937) The role of vitamins in plant development. Bot Rev 3(12):616–640. https://doi.org/10.1007/BF02872294
Bozorg-Haddad O, Azarnivand A, Hosseini-Moghari S-M, Loáiciga HA (2016) Development of a comparative multiple criteria framework for ranking pareto optimal solutions of a multiobjective reservoir operation problem. J Irrig Drain Eng 142(7):e04016019. https://doi.org/10.1061/(ASCE)IR.1943-4774.0001028
Bozorg-Haddad O, Azarnivand A, Hosseini-Moghari S-M, Loáiciga Hugo A (2017) WASPAS application and evolutionary algorithm benchmarking in optimal reservoir optimization problems. J Water Resour Plan Manag 143(1):04016070. https://doi.org/10.1061/(ASCE)WR.1943-5452.0000716
da Silva JAT (2003) Chrysanthemum: advances in tissue culture, cryopreservation, postharvest technology, genetics and transgenic biotechnology. Biotechnol Adv 21(8):715–766. https://doi.org/10.1016/S0734-9750(03)00117-4
da Silva JAT, Kulus D (2014) Chrysanthemum biotechnology: discoveries from the recent literature. Folia Hortic 26(2):67–77. https://doi.org/10.2478/fhort-2014-0007
Demidchik V, Shabala S (2018) Mechanisms of cytosolic calcium elevation in plants: the role of ion channels, calcium extrusion systems and NADPH oxidase-mediated ‘ROS-Ca2+ hub’. Funct Plant Biol 45(2):9–27. https://doi.org/10.1071/FP16420
Dezfooli D, Hosseini-Moghari S-M, Ebrahimi K, Araghinejad S (2018) Classification of water quality status based on minimum quality parameters: application of machine learning techniques. Model Earth Syst Environ 4(1):311–324. https://doi.org/10.1007/s40808-017-0406-9
Ding B, Smith ES, Ding H (2005) Mobilization of the iron centre in IscA for the iron–sulphur cluster assembly in IscU. Biochem J 389(3):797–802. https://doi.org/10.1042/BJ20050405
Dussert S, Verdeil J-L, Buffard-Morel J (1995) Specific nutrient uptake during initiation of somatic embryogenesis in coconut calluses. Plant Sci 111(2):229–236. https://doi.org/10.1016/0168-9452(95)04248-S
Ebrahimian H, Dialameh B, Hosseini-Moghari S-M, Ebrahimian A (2020) Optimal conjunctive use of aqua-agriculture reservoir and irrigation canal for paddy fields (case study: Tajan irrigation network, Iran). Paddy Water Environ 18(3):499–514. https://doi.org/10.1007/s10333-020-00797-5
Elkonin LA, Pakhomova NV (2000) Influence of nitrogen and phosphorus on induction embryogenic callus of sorghum. Plant Cell Tissue Organ Cult 61(2):115–123. https://doi.org/10.1023/A:1006472418218
Etienne H, Lartaud M, Carron M-P, Michaux-Ferrière N (1997) Use of calcium to optimize long-term proliferation of friable embryogenic calluses and plant regeneration in Hevea brasiliensis (Müll. Arg.). J Exp Bot 48(1):129–137. https://doi.org/10.1093/jxb/48.1.129
Farhadi S, Salehi M, Moieni A, Safaie N, Sabet MS (2020) Modeling of paclitaxel biosynthesis elicitation in Corylus avellana cell culture using adaptive neuro-fuzzy inference system-genetic algorithm (ANFIS-GA) and multiple regression methods. PLoS One 15(8):e0237478. https://doi.org/10.1371/journal.pone.0237478
Fisichella M, Silvi E, Morini S (2000) Regeneration of somatic embryos and roots from quince leaves cultured on media with different macroelement composition. Plant Cell Tissue Organ Cult 63(2):101–107. https://doi.org/10.1023/A:1006407803660
Flowers TJ, Colmer TD (2015) Plant salt tolerance: adaptations in halophytes. Ann Bot 115(3):327–331. https://doi.org/10.1093/aob/mcu267
Gago J, Landín M, Gallego PP (2010) Artificial neural networks modeling the in vitro rhizogenesis and acclimatization of Vitis vinifera L. J Plant Physiol 167(15):1226–1231. https://doi.org/10.1016/j.jplph.2010.04.008
Gago J, Martinez-Nunez L, Landin M, Flexas J, Gallego PP (2014) Modeling the effects of light and sucrose on in vitro propagated plants: a multiscale system analysis using artificial intelligence technology. PLoS One 9(1):e85989. https://doi.org/10.1371/journal.pone.0085989
Gago J, Pérez-Tornero O, Landín M, Burgos L, Gallego PP (2011) Improving knowledge of plant tissue culture and media formulation by neurofuzzy logic: a practical case of data mining using apricot databases. J Plant Physiol 168(15):1858–1865. https://doi.org/10.1016/j.jplph.2011.04.008
Galili G, Amir R, Fernie AR (2016) The regulation of essential amino acid synthesis and accumulation in plants. Annu Rev Plant Biol 67:153–178. https://doi.org/10.1146/annurev-arplant-043015-112213
Gamborg OLC, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50(1):151–158. https://doi.org/10.1016/0014-4827(68)90403-5
García-Pérez P, Lozano-Milo E, Landín M, Gallego PP (2020a) Combining medicinal plant in vitro culture with machine learning technologies for maximizing the production of phenolic compounds. Antioxidants 9(3):210. https://doi.org/10.3390/antiox9030210
García-Pérez P, Lozano-Milo E, Landín M, Gallego PP (2020b) Machine learning technology reveals the concealed interactions of phytohormones on medicinal plant in vitro organogenesis. Biomolecules 10(5):746. https://doi.org/10.3390/biom10050746
Ghosh B, Sen S (1991) Plant regeneration through somatic embryogenesis from spear callus culture of Asparagus cooperi Baker. Plant Cell Rep 9(12):667–670. https://doi.org/10.1007/BF00235353
Greenway MB, Phillips IC, Lloyd MN, Hubstenberger JF, Phillips GC (2012) A nutrient medium for diverse applications and tissue growth of plant species in vitro. Vitro Cell Dev Biol Plant 48(4):403–410. https://doi.org/10.1007/s11627-012-9452-1
Greer MS, Kovalchuk I, Eudes F (2009) Ammonium nitrate improves direct somatic embryogenesis and biolistic transformation of Triticum aestivum. New Biotechnol 26(1–2):44–52. https://doi.org/10.1016/j.nbt.2009.02.003
Groll J, Mycock D, Gray V (2002) Effect of medium salt concentration on differentiation and maturation of somatic embryos of cassava (Manihot esculenta Crantz). Ann Bot 89(5):645–648. https://doi.org/10.1093/aob/mcf095
Guillou C, Fillodeau A, Brulard E, Breton D, Maraschin SDF, Verdier D, Simon M, Ducos J-P (2018) Indirect somatic embryogenesis of Theobroma cacao L. in liquid medium and improvement of embryo-to-plantlet conversion rate. In Vitro Cell Dev Biol Plant 54(4):377–391. https://doi.org/10.1007/s11627-018-9909-y
Haddad Omid B, Hosseini-Moghari S-M, Loáiciga Hugo A (2016) Biogeography-based optimization algorithm for optimal operation of reservoir systems. J Water Resour Plan Manag 142(1):04015034. https://doi.org/10.1061/(ASCE)WR.1943-5452.0000558
Hararuk O, Zwart JA, Jones SE, Prairie Y, Solomon CT (2018) Model-data fusion to test hypothesized drivers of lake carbon cycling reveals importance of physical controls. J Geophys Res-Biogeo 123(3):1130–1142. https://doi.org/10.1002/2017JG004084
Hesami M, Alizadeh M, Naderi R, Tohidfar M (2020a) Forecasting and optimizing Agrobacterium-mediated genetic transformation via ensemble model- fruit fly optimization algorithm: a data mining approach using chrysanthemum databases. PLoS One 15:e0239901. https://doi.org/10.1371/journal.pone.0239901
Hesami M, Condori-Apfata JA, Valderrama Valencia M, Mohammadi M (2020b) Application of artificial neural network for modeling and studying in vitro genotype-independent shoot regeneration in wheat. Appl Sci 10(15):5370. https://doi.org/10.3390/app10155370
Hesami M, Daneshvar MH (2018a) In vitro adventitious shoot regeneration through direct and indirect organogenesis from seedling-derived hypocotyl segments of Ficus religiosa L.: an important medicinal plant. HortScience 53(1):55–61. https://doi.org/10.21273/HORTSCI12637-17
Hesami M, Daneshvar MH (2018b) Indirect organogenesis through seedling-derived leaf segments of Ficus religiosa-a multipurpose woody medicinal plant. J Crop Sci Biotechnol 21(2):129–136. https://doi.org/10.1007/s12892-018-0024-0
Hesami M, Daneshvar MH, Lotfi A (2017a) In vitro shoot proliferation through cotyledonary node and shoot tip explants of Ficus religiosa L. Plant Tissue Cult Biotechnol 27(1):85–88. https://doi.org/10.3329/ptcb.v27i1.35017
Hesami M, Daneshvar MH, Yoosefzadeh-Najafabadi M (2018a) Establishment of a protocol for in vitro seed germination and callus formation of Ficus religiosa L., an important medicinal plant. Jundishapur J Nat Pharm Prod 13(4):e62682. https://doi.org/10.5812/jjnpp.62682
Hesami M, Daneshvar MH, Yoosefzadeh-Najafabadi M (2019a) An efficient in vitro shoot regeneration through direct organogenesis from seedling-derived petiole and leaf segments and acclimatization of Ficus religiosa. J For Res 30(3):807–815
Hesami M, Jones AMP (2020) Application of artificial intelligence models and optimization algorithms in plant cell and tissue culture. Appl Microbiol Biotechnol 104:1–37. https://doi.org/10.1007/s00253-020-10888-2
Hesami M, Naderi R, Tohidfar M (2019b) Modeling and optimizing in vitro sterilization of chrysanthemum via multilayer perceptron-non-dominated sorting genetic algorithm-II (MLP-NSGAII). Front Plant Sci 10:282. https://doi.org/10.3389/fpls.2019.00282
Hesami M, Naderi R, Tohidfar M (2019c) Modeling and optimizing medium composition for shoot regeneration of chrysanthemum via radial basis function-non-dominated sorting genetic algorithm-II (RBF-NSGAII). Sci Rep 9(1):1–11. https://doi.org/10.1038/s41598-019-54257-0
Hesami M, Naderi R, Tohidfar M, Yoosefzadeh-Najafabadi M (2019d) Application of adaptive neuro-fuzzy inference system-non-dominated sorting genetic algorithm-II (ANFIS-NSGAII) for modeling and optimizing somatic embryogenesis of chrysanthemum. Front Plant Sci 10:869. https://doi.org/10.3389/fpls.2019.00869
Hesami M, Naderi R, Tohidfar M, Yoosefzadeh-Najafabadi M (2020c) Development of support vector machine-based model and comparative analysis with artificial neural network for modeling the plant tissue culture procedures: effect of plant growth regulators on somatic embryogenesis of chrysanthemum, as a case study. Plant Methods 16(1):112. https://doi.org/10.1186/s13007-020-00655-9
Hesami M, Naderi R, Yoosefzadeh-Najafabadi M (2018b) Optimizing sterilization conditions and growth regulator effects on in vitro shoot regeneration through direct organogenesis in Chenopodium quinoa. BioTechnologia 99(1):49–57. https://doi.org/10.5114/bta.2018.73561
Hesami M, Naderi R, Yoosefzadeh-Najafabadi M, Maleki M (2018c) In vitro culture as a powerful method for conserving Iranian ornamental geophytes. BioTechnologia 99(1):73–81. https://doi.org/10.5114/bta.2018.73563
Hesami M, Naderi R, Yoosefzadeh-Najafabadi M, Rahmati M (2017b) Data-driven modeling in plant tissue culture. J Appl Environ Biol Sci 7(8):37–44
Hildebrandt AC, Riker A, Duggar B (1946) The influence of the composition of the medium on growth in vitro of excised tobacco and sunflower tissue cultures. Am J Bot 33:591–597. https://doi.org/10.2307/2437399
Hosseini-Moghari S-M, Araghinejad S, Azarnivand A (2017) Drought forecasting using data-driven methods and an evolutionary algorithm. Model Earth Syst Environ 3(4):1675–1689. https://doi.org/10.1007/s40808-017-0385-x
Hosseini-Moghari S-M, Morovati R, Moghadas M, Araghinejad S (2015) Optimum operation of reservoir using two evolutionary algorithms: imperialist competitive algorithm (ICA) and cuckoo optimization algorithm (COA). Water Resour Manag 29(10):3749–3769. https://doi.org/10.1007/s11269-015-1027-6
Hosseini-Moghari SM, Araghinejad S (2015) Monthly and seasonal drought forecasting using statistical neural networks. Environ Earth Sci 74(1):397–412. https://doi.org/10.1007/s12665-015-4047-x
Jamshidi S, Yadollahi A, Arab MM, Soltani M, Eftekhari M, Sabzalipoor H, Sheikhi A, Shiri J (2019) Combining gene expression programming and genetic algorithm as a powerful hybrid modeling approach for pear rootstocks tissue culture media formulation. Plant Methods 15(1):136. https://doi.org/10.1186/s13007-019-0520-y
Jansen MA, Booij H, Schel JH, de Vries SC (1990) Calcium increases the yield of somatic embryos in carrot embryogenic suspension cultures. Plant Cell Rep 9(4):221–223. https://doi.org/10.1007/BF00232184
Jones B, Gunnerås SA, Petersson SV, Tarkowski P, Graham N, May S, Dolezal K, Sandberg G, Ljung K (2010) Cytokinin regulation of auxin synthesis in Arabidopsis involves a homeostatic feedback loop regulated via auxin and cytokinin signal transduction. Plant Cell 22:2956–2969. https://doi.org/10.1105/tpc.110.074856
Khvatkov P, Chernobrovkina M, Okuneva A, Dolgov S (2019) Creation of culture media for efficient duckweeds micropropagation (Wolffia arrhiza and Lemna minor) using artificial mathematical optimization models. Plant Cell Tissue Organ Cult 136(1):85–100. https://doi.org/10.1007/s11240-018-1494-
Kim HJ, Lee YS (2005) Identification of new dicaffeoylquinic acids from Chrysanthemum morifolium and their antioxidant activities. Planta Med 71(09):871–876. https://doi.org/10.1055/s-2005-873115
Kintzios S, Drossopoulos J, Lymperopoulos C (2001) Effect of vitamins and inorganic micronutrients on callus growth and somatic embryogenesis from leaves of chilli pepper. Plant Cell Tissue Organ Cult 67(1):55–62. https://doi.org/10.1023/A:1011610413177
Kintzios S, Stavropoulou E, Skamneli S (2004) Accumulation of selected macronutrients and carbohydrates in melon tissue cultures: association with pathways of in vitro dedifferentiation and differentiation (organogenesis, somatic embryogenesis). Plant Sci 167(3):655–664. https://doi.org/10.1016/j.plantsci.2004.05.021
Knutsson P, Cantatore V, Seemann M, Tam PL, Panas I (2018) Role of potassium in the enhancement of the catalytic activity of calcium oxide towards tar reduction. Appl Catal B-Environ 229:88–95. https://doi.org/10.1016/j.apcatb.2018.02.002
Koleva-Gudeva LR, Spasenoski M, Trajkova F (2007) Somatic embryogenesis in pepper anther culture: the effect of incubation treatments and different media. Sci Hortic 111(2):114–119. https://doi.org/10.1016/j.scienta.2006.10.013
Kothari S, Agarwal K, Kumar S (2004) Inorganic nutrient manipulation for highly improved in vitro plant regeneration in finger millet—Eleusine coracana (L.) Gaertn. In Vitro Cell Dev Biol Plant 40(5):515–519. https://doi.org/10.1079/IVP2004564
Lin G-H, Lin L, Liang H-W, Ma X, Wang J-Y, Wu L-P, Jiang H-D, Bruce IC, Xia Q (2010) Antioxidant action of a Chrysanthemum morifolium extract protects rat brain against ischemia and reperfusion injury. J Med Food 13(2):306–311. https://doi.org/10.1089/jmf.2009.1184
Loh C, Lim G (1992) The influence of medium components on secondary embryogenesis of winter oilseed rape, Brassica napus L. ssp. oleifera (Metzg.) Sink. New Phytol 121(3):425–430. https://doi.org/10.1111/j.1469-8137.1992.tb02942.x
Luo J-P, Jiang S-T, Pan L-J (2003) Cold-enhanced somatic embryogenesis in cell suspension cultures of Astragalus adsurgens Pall.: relationship with exogenous calcium during cold pretreatment. Plant Growth Regul 40(2):171–177. https://doi.org/10.1023/A:1024295901808
May R, Trigiano R (1991) Somatic embryogenesis and plant regeneration from leaves of Dendranthema grandiflora. J Am Soc Hortic Sci 116(2):366–371
Milazzo MC, Zheng Z, Kellett G, Haynesworth K, Shetty K (1999) Stimulation of benzyladenine-induced in vitro shoot organogenesis and endogenous proline in melon (Cucumis melo L.) by fish protein hydrolysates in combination with proline analogues. J Agric Food Chem 47(4):1771–1775. https://doi.org/10.1021/jf9812883
Minhas AP, Tuli R, Puri S (2018) Pathway editing targets for thiamine biofortification in rice grains. Front Plant Sci 9:e975. https://doi.org/10.3389/fpls.2018.00975
Minyaka E, Niemenak N, Sangare A, Omokolo DN (2008) Effect of MgSO4 and K2SO4 on somatic embryo differentiation in Theobroma cacao L. Plant Cell Tissue Organ Cult 94(2):149–160. https://doi.org/10.1007/s11240-008-9398-5
Moravej M, Amani P, Hosseini-Moghari S-M (2020) Groundwater level simulation and forecasting using interior search algorithm-least square support vector regression (ISA-LSSVR). Groundw Sustain Dev 11:1–18
Moravej M, Hosseini-Moghari S-M (2016) Large scale reservoirs system operation optimization: the interior search algorithm (ISA) approach. Water Resour Manag 30(10):3389–3407. https://doi.org/10.1007/s11269-016-1358-y
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15(3):473–497. https://doi.org/10.1111/j.1399-3054.1962.tb08052.x
Naing AH, Kim CK, Yun BJ, Jin JY, Lim KB (2013) Primary and secondary somatic embryogenesis in Chrysanthemum cv. Euro Plant Cell Tiss Org Cult 112(3):361–368. https://doi.org/10.1007/s11240-012-0243-5
Nezami-Alanagh E, Garoosi G-A, Maleki S, Landín M, Gallego PP (2017) Predicting optimal in vitro culture medium for Pistacia vera micropropagation using neural networks models. Plant Cell Tissue Organ Cult 129(1):19–33. https://doi.org/10.1007/s11240-016-1152-9
Niazian M, Sadat-Noori SA, Abdipour M, Tohidfar M, Mortazavian SMM (2018) Image processing and artificial neural network-based models to measure and predict physical properties of embryogenic callus and number of somatic embryos in ajowan (Trachyspermum ammi (L.) Sprague). In Vitro Cell Dev-Pl 54(1):54–68. https://doi.org/10.1007/s11627-017-9877-7
Niedz RP, Evens TJ (2008) The effects of nitrogen and potassium nutrition on the growth of nonembryogenic and embryogenic tissue of sweet orange (Citrus sinensis (L.) Osbeck). BMC Plant Biol 8(1):126. https://doi.org/10.1186/1471-2229-8-126
Nitsch JP, Nitsch C (1969) Haploid plants from pollen grains. Science 163(3862):85–87. https://doi.org/10.1126/science.163.3862.85
Noda N, Yoshioka S, Kishimoto S, Nakayama M, Douzono M, Tanaka Y, Aida R (2017) Generation of blue chrysanthemums by anthocyanin B-ring hydroxylation and glucosylation and its coloration mechanism. Sci Adv 3(7):e1602785. https://doi.org/10.1126/sciadv.1602785
Norton M, LaBonte D (1996) Cultural conditions affecting the frequency of embryoid formation in sweetpotato (Ipomoea batatas). HortScience 31(4):630–630
Pang J, Ryan MH, Lambers H, Siddique KH (2018) Phosphorus acquisition and utilisation in crop legumes under global change. Curr Opin Plant Biol 45:248–254. https://doi.org/10.1016/j.pbi.2018.05.012
Sa N, Rawat R, Thornburg C, Walker KD, Roje S (2016) Identification and characterization of the missing phosphatase on the riboflavin biosynthesis pathway in Arabidopsis thaliana. Plant J 88(5):705–716. https://doi.org/10.1111/tpj.13291
Saha P, Bandyopadhyay S, Raychaudhuri SS (2011) Formulation of nutrient medium for in vitro somatic embryo induction in Plantago ovata Forsk. Biol Trace Elem Res 140(2):225–243. https://doi.org/10.1007/s12011-010-8684-3
Saito K (2004) Sulfur assimilatory metabolism. The long and smelling road. Plant Physiol 136(1):2443–2450. https://doi.org/10.1104/pp.104.046755
Salehi M, Farhadi S, Moieni A, Safaie N, Ahmadi H (2020) Mathematical modeling of growth and paclitaxel biosynthesis in Corylus avellana cell culture responding to fungal elicitors using multilayer perceptron-genetic algorithm. Front Plant Sci 11:1148. https://doi.org/10.3389/fpls.2020.01148
Samson NP, Campa C, Le Gal L, Noirot M, Thomas G, Lokeswari T, De Kochko A (2006) Effect of primary culture medium composition on high frequency somatic embryogenesis in different Coffea species. Plant Cell Tissue Organ Cult 86(1):37–45. https://doi.org/10.1007/s11240-006-9094-2
Schenk RU, Hildebrandt A (1972) Medium and techniques for induction and growth of monocotyledonous and dicotyledonous plant cell cultures. Can J Bot 50(1):199–204. https://doi.org/10.1139/b72-026
Selby C, Harvey B (1990) The influence of composition of the basal medium on the growth and morphogenesis of cultured Sitka spruce (Picea sitchensis) tissues. Ann Bot 65(4):395–407. https://doi.org/10.1093/oxfordjournals.aob.a087950
Sheikhi A, Mirdehghan SH, Arab MM, Eftekhari M, Ahmadi H, Jamshidi S, Gheysarbigi S (2020) Novel organic-based postharvest sanitizer formulation using Box Behnken design and mathematical modeling approach: a case study of fresh pistachio storage under modified atmosphere packaging. Postharvest Biol Technol 160:111047. https://doi.org/10.1016/j.postharvbio.2019.111047
Shetty K, McKersie BD (1993) Proline, thioproline and potassium mediated stimulation of somatic embryogenesis in alfalfa (Medicago sativa L.). Plant Sci 88(2):185–193. https://doi.org/10.1016/0168-9452(93)90090-M
Smith AD, Agar JN, Johnson KA, Frazzon J, Amster IJ, Dean DR, Johnson MK (2001) Sulfur transfer from IscS to IscU: the first step in iron− sulfur cluster biosynthesis. J Am Chem Soc 123(44):11103–11104. https://doi.org/10.1021/ja016757n
Tanaka K, Kanno Y, Kudo S, Suzuki M (2000) Somatic embryogenesis and plant regeneration in chrysanthemum (Dendranthema grandiflorum (Ramat.) Kitamura). Plant Cell Rep 19(10):946–953. https://doi.org/10.1007/s002990000225
Taylor NJ, Edwards M, Kiernan RJ, Davey CD, Blakesley D, Henshaw GG (1996) Development of friable embryogenic callus and embryogenic suspension culture systems in cassava (Manihot esculenta Crantz). Nat Biotechnol 14(6):726–730. https://doi.org/10.1038/nbt0696-726
Trigiano R, May R, Conger B (1992) Reduced nitrogen influences somatic embryo quality and plant regeneration from suspension cultures of orchardgrass. Vitro Cell Dev Biol Plant 28(4):187–191. https://doi.org/10.1007/BF02823315
Walker K, Sato S (1981) Morphogenesis in callus tissue of Medicago sativa: the role of ammonium ion in somatic embryogenesis. Plant Cell Tissue Organ Cult 1(1):109–121. https://doi.org/10.1007/BF02318910
White PR (1942) Plant tissue cultures. Annu Rev Biochem 11(1):615–628. https://doi.org/10.1146/annurev.bi.11.070142.003151
Wu X-M, Zhang Q-Z, Wang Y-Z (2018) Traceability of wild Paris polyphylla Smith var. yunnanensis based on data fusion strategy of FT-MIR and UV–Vis combined with SVM and random forest. Spectrochim Acta A Mol Biomol Spectrosc 205:479–488. https://doi.org/10.1016/j.saa.2018.07.067
Xu P, Zhang Z, Wang B, Xia X, Jia J (2012) Somatic embryogenesis and plant regeneration in chrysanthemum (Yuukou). Plant Cell Tissue Organ Cult 111(3):393–397. https://doi.org/10.1007/s11240-012-0201-2
Xu XM, Møller SG (2004) AtNAP7 is a plastidic SufC-like ATP-binding cassette/ATPase essential for Arabidopsis embryogenesis. Proc Natl Acad Sci 101(24):9143–9148. https://doi.org/10.1073/pnas.0400799101
Yan Y-W, Mao D-D, Yang L, Qi J-L, Zhang X-X, Tang Q-L, Li Y-P, Tang R-J, Luan S (2018) Magnesium transporter MGT6 plays an essential role in maintaining magnesium homeostasis and regulating high magnesium tolerance in Arabidopsis. Front Plant Sci 9:e274. https://doi.org/10.3389/fpls.2018.00274
Yang L, Aobulikasimu N, Cheng P, Wang J-H, Li H (2017) Analysis of floral volatile components and antioxidant activity of different varieties of Chrysanthemum morifolium. Molecules 22(10):1790. https://doi.org/10.3390/molecules22101790
Zhao Y (2008) The role of local biosynthesis of auxin and cytokinin in plant development. Curr Opin Plant Biol 11(1):16–22. https://doi.org/10.1016/j.pbi.2007.10.008
Author information
Authors and Affiliations
Contributions
M.H. was responsible for performing the experiments, data modeling, summing up, and writing the manuscript. R.N. and M.T. were responsible for designing and leading the experiments and revising the manuscript. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights
This work does not involve any human participation nor live animals performed by any of the listed authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(PDF 567 kb)
Rights and permissions
About this article
Cite this article
Hesami, M., Naderi, R. & Tohidfar, M. Introducing a hybrid artificial intelligence method for high-throughput modeling and optimizing plant tissue culture processes: the establishment of a new embryogenesis medium for chrysanthemum, as a case study. Appl Microbiol Biotechnol 104, 10249–10263 (2020). https://doi.org/10.1007/s00253-020-10978-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10978-1