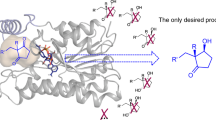
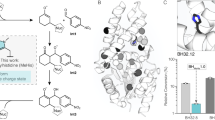
Abstract
ChKRED20 is an efficient and robust anti-Prelog ketoreductase that can catalyze the reduction of ketones to chiral alcohols as pharmaceutical intermediates with great industrial potential. To overcome its limitation on the bioreduction of ortho-substituted acetophenone derivatives, the X-ray crystal structure of the apo-enzyme of ChKRED20 was determined at a resolution of 1.85 Å and applied to the molecular modeling and reshaping of the catalytic cavity via three rounds of iterative saturation mutagenesis together with alanine scanning and recombination. The mutant Mut3B was achieved with expanded catalytic scope that covered all the nine substrates tested as compared with two substrates for the wild type. It exhibited 13–20-fold elevated k cat/K m values relative to the wild type or to the first gain-of-activity mutant, while retaining excellent stereoselectivity toward seven of the substrates (98–> 99% ee). Another mutant 29G10 displayed complementary selectivity for eight of the ortho-substituted acetophenone derivatives, with six of them delivering excellent stereoselectivity (90–99% ee). Its k cat/K m value toward 1-(2-fluorophenyl)ethanone was 5.6-fold of the wild type. The application of Mut3B in elevated substrate concentrations of 50–100 g/l was demonstrated in 50-ml reactions, achieving 75–> 99% conversion and > 99% ee.






Similar content being viewed by others
References
Acevedo-Rocha CG, Hoebenreich S, Reetz MT (2014) Iterative saturation mutagenesis: a powerful approach to engineer proteins by systematically simulating Darwinian evolution. Methods Mol Bio 1179:103–128
Bailey S (1994) The CCP4 suite—programs for protein crystallography. Acta Crystallogr D 50:760–763
Bornscheuer UT, Huisman GW, Kazlauskas RJ, Lutz S, Moore JC, Robins K (2012) Engineering the third wave of biocatalysis. Nature 485(7397):185–194
Busing I, Hoffken HW, Breuer M, Wohlbrand L, Hauer B, Rabus R (2015) Molecular genetic and crystal structural analysis of 1-(4-hydroxyphenyl)-ethanol dehydrogenase from ‘Aromatoleum aromaticum’ EbN1. J Mol Microbiol Biotechnol 25(5):327–339
Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D 66:12–21
Chovancova E, Pavelka A, Benes P, Strnad O, Brezovsky J, Kozlikova B, Gora A, Sustr V, Klvana M, Medek P (2012) CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput Biol 8(10):e1002708
Despina BJ, Kille S, Taglieber A, Reetz MT (2009) Directed evolution of an enantioselective enoate-reductase: testing the utility of iterative saturation mutagenesis. Adv Synth Catal 351(18):3287–3305
Duax WL, Huether R, Pletnev V, Umland TC, Weeks CM (2009) Divergent evolution of a Rossmann fold and identification of its oldest surviving ancestor. Int J Bioinform Res App 5:280–294
Dudzik A, Snoch W, Borowiecki P, Opalinska-Piskorz J, Witko M, Heider J, Szaleniec M (2015) Asymmetric reduction of ketones and beta-keto esters by (S)-1-phenylethanol dehydrogenase from denitrifying bacterium Aromatoleum aromaticum. Appl Microbiol Biotechnol 99(12):5055–5069
Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res 34:116–118
Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D 60:2126–2132
Filling C, Berndt KD, Benach J, Knapp S, Prozorovski T, Nordling E, Ladenstein R, Jörnvall H, Oppermann U (2002) Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J Biol Chem 277(28):25677–25684
Glieder A, Farinas ET, Arnold FH (2002) Laboratory evolution of a soluble, self-sufficient, highly active alkane hydroxylase. Nat Biotechnol 20(11):1135–1139
Gooding OW, Voladri R, Bautista A, Hopkins T, Huisman G, Jenne S, Ma S, Mundorff EC, Savile MM (2010) Development of a practical biocatalytic process for (R)-2-methylpentanol. Org Process Res Dev 14(1):119–126
Hollmann F, Arends IWCE, Holtmann D (2011) Enzymatic reductions for the chemist. Green Chem 13(9):2285–2314
Holm L, Laakso LM (2016) DALI server update. Nucleic Acids Res 44(W1):W351–W355
Huisman GW, Collier SJ (2013) On the development of new biocatalytic processes for practical pharmaceutical synthesis. Curr Opin Chem Biol 17(2):284–292
Huisman GW, Liang J, Krebber A (2010) Practical chiral alcohol manufacture using ketoreductases. Curr Opin Chem Biol 14(2):122–129
Hummel W (1997) New alcohol dehydrogenases for the synthesis of chiral compounds. Adv Biochem Eng Biotechnol 58:145–184
Jornvall H, Persson B, Krook M, Atrian S, Gonzalezduarte R, Jeffery J, Ghosh D (1995) Short-chain dehydrogenases reductases (SDR). Biochemistry 34(18):6003–6013
Kille S, Acevedorocha CG, Parra LP, Zhang ZG, Opperman DJ, Reetz MT, Acevedo JP (2013) Reducing codon redundancy and screening effort of combinatorial protein libraries created by saturation mutagenesis. ACS Synth Biol 2(2):83–92
Lewis JC, Mantovani SM, Fu Y, Snow CD, Komor RS, Wong CH, Arnold FH (2010) Combinatorial alanine substitution enables rapid optimization of cytochrome P450(BM3) for selective hydroxylation of large substrates. Chembiochem 11(18):2502–2505
Li AP, Ye LD, Yang XH, Yang CC, Gu JL, Yu HW (2016) Structure-guided stereoselectivity inversion of a short-chain dehydrogenase/reductase towards halogenated acetophenones. Chem Commun 52(37):6284–6287
Li C, Liu Y, Pei X-Q, Wu Z-L (2017a) Stereo-complementary bioreduction of saturated N-heterocyclic ketones. Process Biochem 56:90–97
Li M, Zhang ZJ, Kong XD, HL Y, Zhou J, Xu JH (2017b) Engineering Streptomyces coelicolor carbonyl reductase for efficient synthesis of atorvastatin precursor. Appl Environ Microbiol 83(12):e00603–e00617
Liang J, Mundorff E, Voladri R, Jenne S, Gilson L, Conway A, Krebber A, Wong J, Huisman G, Truesdell S, Lalonde J (2010) Highly enantioselective reduction of a small heterocyclic ketone: Biocatalytic reduction of tetrahydrothiophene-3-one to the corresponding (R)-alcohol. Org Process Res Dev 14(1):188–192
Liu Y, Tang T-X, Pei X-Q, Zhang C, Wu Z-L (2014) Identification of ketone reductase ChKRED20 from the genome of Chryseobacterium sp. CA49 for highly efficient anti-Prelog reduction of 3,5-bis(trifluoromethyl)acetophenone. J Mol Catal B Enzym 102:1–8
Liu Y-C, Guo C, Liu Y, Wang H-B, Wu ZL (2017) Enzymatic cascades for the stereo-complementary epimerisation of in situ generated epoxy alcohols. Org Biomol Chem 15(12):2562–2568
Martins BM, Macedo-Ribeiro S, Bresser J, Buckel W, Messerschmidt A (2005) Structural basis for stereo-specific catalysis in NAD+-dependent (R)-2-hydroxyglutarate dehydrogenase from Acidaminococcus fermentans. FEBS J 272(1):269–281
McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40:658–674
Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53:240–255
Nealon CM, Musa MM, Patel JM, Phillips RS (2015) Controlling substrate specificity and stereospecificity of alcohol dehydrogenases. ACS Catal 5(4):2100–2114
Noey EL, Tibrewal N, Jimenez-Oses G, Osuna S, Park J, Bond CM, Cascio D, Liang J, Zhang XY, Huisman GW, Tang Y, Houk KN (2015) Origins of stereoselectivity in evolved ketoreductases. P Natl Acad Sci USA 112(51):E7065–E7072
Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol 276:307–326
Pennacchio A, Sannino V, Sorrentino G, Rossi M, Raia CA, Esposito L (2013) Biochemical and structural characterization of recombinant short-chain NAD(H)-dependent dehydrogenase/reductase from Sulfolobus acidocaldarius highly enantioselective on diaryl diketone benzil. Appl Microbiol Biotechnol 97(9):3949–3964
Prelog V (2009) Specification of the stereospecificity of some oxido-reductases by diamond lattice sections. Pure Appl Chem 9(1):119–130
Reetz MT, Prasad S, Carballeira JD, Gumulya Y, Bocola M (2010) Iterative saturation mutagenesis accelerates laboratory evolution of enzyme stereoselectivity: Rigorous comparison with traditional methods. J Am Chem Soc 132(26):9144–9152
Schlieben NH, Niefind K, Muller J, Riebel B, Hummel W, Schomburg D (2005) Atomic resolution structures of R-specific alcohol dehydrogenase from Lactobacillus brevis provide the structural bases of its substrate and cosubstrate specificity. J Mol Biol 349(4):801–813
Shang YP, Chen Q, Kong XD, Zhang YJ, Xu JH, Yu HL (2017) Efficient synthesis of (R)-2-chloro-1-(2,4-dichlorophenyl)ethanol with a ketoreductase from Scheffersomyces stipitis CBS 6045. Adv Synth Catal 359(3):426–431
Sun ZT, Lonsdale R, Ilie A, Li GY, Zhou JH, Reetz MT (2016) Catalytic asymmetric reduction of difficult-to-reduce ketones: triple-code saturation mutagenesis of an alcohol dehydrogenase. ACS Catal 6(3):1598–1605
Tang T-X, Liu Y, Wu Z-L (2014) Characterization of a robust anti-Prelog short-chain dehydrogenase/reductase ChKRED20 from Chryseobacterium sp. CA49. J Mol Catal B Enzym 105:82–88
Träff A, Bogár K, Warner M, Bäckvall J-E (2008) Highly efficient route for enantioselective preparation of chlorohydrins via dynamic kinetic resolution. Org Lett 10(21):4807–4810
Wang LJ, Li CX, Ni Y, Zhang J, Liu X, Xu JH (2011) Highly efficient synthesis of chiral alcohols with a novel NADH-dependent reductase from Streptomyces coelicolor. Bioresour Technol 102(14):7023–7028
Wang S, Nie Y, Xu Y, Zhang R, Ko TP, Huang CH, Chan HC, Guo RT, Xiao R (2014) Unconserved substrate-binding sites direct the stereoselectivity of medium-chain alcohol dehydrogenase. Chem Commun 50(58):7770–7772
Weckbecker A, Hummel W (2006) Cloning, expression, and characterization of an (R)-specific alcohol dehydrogenase from Lactobacillus kefir. Biocatal Biotransform 24(5):380–389
Zhang D, Chen X, Chi J, Feng J, Wu Q, Zhu D (2015a) Semi-rational engineering a carbonyl reductase for the enantioselective reduction of β-amino ketones. ACS Catal 5(4):2452–2457
Zhang R, Xu Y, Xiao R (2015b) Redesigning alcohol dehydrogenases/reductases for more efficient biosynthesis of enantiopure isomers. Biotechnol Adv 33(8):1671–1684
Zhang W, Tang WL, Wang DIC, Li Z (2011) Concurrent oxidations with tandem biocatalysts in one pot: green, selective and clean oxidations of methylene groups to ketones. Chem Commun 47(11):3284–3286
Zhao F-J, Liu Y, Pei X-Q, Guo C, Wu Z-L (2017) Single mutations of ketoreductase ChKRED20 enhance the bioreductive production of (1S)-2-chloro-1-(3, 4-difluorophenyl) ethanol. Appl Microbiol Biotechnol 101(5):1945–1952
Zhao F-J, Pei X-Q, Ren Z-Q, Wu Z-L (2016) Rapid asymmetric reduction of ethyl 4-chloro-3-oxobutanoate using a thermostabilized mutant of ketoreductase ChKRED20. Appl Microbiol Biotechnol 100(8):3567–3575
Zheng YG, Yin HH, DF Y, Chen X, Tang XL, Zhang XJ, Xue YP, Wang YJ, Liu ZQ (2017) Recent advances in biotechnological applications of alcohol dehydrogenases. Appl Microbiol Biotechnol 101(3):987–1001
Acknowledgements
Shanghai Synchrotron Radiation Facilities (SSRF) and National Center for Protein Science Shanghai (NCPSS), China are gratefully acknowledged for the provision of synchrotron radiation facilities and efficient support.
Funding
This study was funded by the National Natural Science Foundation of China (21372216, 21572220, and 21708038).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 1188 kb)
Rights and permissions
About this article
Cite this article
Zhao, FJ., Jin, Y., Liu, Z. et al. Crystal structure and iterative saturation mutagenesis of ChKRED20 for expanded catalytic scope. Appl Microbiol Biotechnol 101, 8395–8404 (2017). https://doi.org/10.1007/s00253-017-8556-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8556-2