Abstract
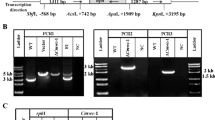
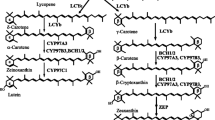
Cryptochromes (CRYs) belong to the photolyase/cryptochrome flavoprotein family, which is widely distributed in all kingdoms. A phylogenetic analysis indicated that three Cordyceps militaris proteins [i.e., cryptochrome DASH (CmCRY-DASH), (6-4) photolyase, and cyclobutane pyrimidine dimer (CPD) class I photolyase] belong to separate fungal photolyase/cryptochrome subfamilies. CmCRY-DASH consists of DNA photolyase and flavin adenine dinucleotide-binding domains, with RGG repeats in a C-terminal extension. Considerably, more carotenoids and cordycepin accumulated in the ΔCmcry-DASH strain than in the wild-type or ΔCmwc-1 strains, indicating an inhibitory role for CmCRY-DASH in these biosynthetic pathways. Fruiting body primordia could form in the ΔCmcry-DASH strain, but the fruiting bodies were unable to elongate normally, differently from the Cmwc-1 disruption strain, where primordium differentiation did not occur. Cmcry-DASH expression is induced by light in the wild-type strain, but not in the ΔCmwc-1 strain. CmCRY-DASH is also necessary for the expression of Cmwc-1, implying that Cmcry-DASH and Cmwc-1 exhibit interdependent expression. The Cmvvd expression levels in the wild-type and ΔCmcry-DASH strains increased considerably following irradiation, while Cmvvd expression in the ΔCmwc-1 strain was not induced by light. It is speculated that the photo adaptation may be faster in the Cmcry-DASH mutant based on Cmvvd transcript dynamics. These results provide new insights into the biological functions of fungal DASH CRYs. Furthermore, the DASH CRYs may regulate fruiting body development and secondary metabolism differently than WC-1.







Similar content being viewed by others
References
Bayram Ö, Biesemann C, Krappmann S, Galland P, Braus GH (2008) More than a repair enzyme: Aspergillus nidulans photolyase-like CryA is a regulator of sexual development. Mol Biol Cell 19(8):3254–3262
Bayram Ö, Braus GH, Fischer R, Rodriguez-Romero J (2010) Spotlight on Aspergillus nidulans photosensory systems. Fungal Genet Biol 47(11):900–908
Brudler R, Hitomi K, Daiyasu H, Toh H, Kucho K, Ishiura M, Kanehisa M, Roberts AV, Todo T, Tainer JA, Getzoff ED (2003) Identification of a new cryptochrome class: structure, function, and evolution. Mol Cell 11(1):59–67
Cashmore AR, Jarillo JA, Wu Y, Liu D (1999) Cryptochromes: blue light receptors for plants and animals. Science 284(5415):760–765
Castrillo M, Avalos J (2015) The flavoproteins CryD and VvdA cooperate with the white collar protein WcoA in the control of photocarotenogenesis in Fusarium fujikuroi. PLoS One 10(3):e0119785
Castrillo M, García-Martínez J, Avalos J (2013) Light-dependent functions of the Fusarium fujikuroi CryD DASH cryptochrome in development and secondary metabolism. Appl Environ Microbiol 79(8):2777–2788
Chang ST, Hayes WA (1978) The biology and cultivation of edible mushrooms. Academic Press, New York
Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GT, Batschauer A, Ahmad M (2011) The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol 62:335–364
Chen CH, DeMay BS, Gladfelter AS, Dunlap JC, Loros JJ (2010) Physical interaction between VIVID and white collar complex regulates photoadaptation in Neurospora. Proc Natl Acad Sci U S A 107:16715–16720
Daiyasu H, Ishikawa T, Kuma K, Iwai S, Todo T, Toh H (2004) Identification of cryptochrome DASH from vertebrates. Genes Cells 9(5):479–495
Dasgupta A, Fuller KK, Dunlap JC, Loro JJ (2016) Seeing the world differently: variability in the photosensory mechanisms of two model fungi. Environ Microbiol 18(1):5–20
Dong CH, Yao YJ (2010) Comparison of some metabolites among cultured mycelia of Ophiocordyceps sinensis from different geographical regions. Int J Med Mushrooms 12:287–297
Dong CH, Li WJ, Li ZZ, Yan WJ, Li TH, Liu XZ (2016) Cordyceps industry in China: current status, challenges and perspectives—Jinhu declaration for cordyceps industry development. Mycosystema 35(1):1–15 (in Chinese)
Fang W, St Leger RJ (2010) Mrt, a gene unique to fungi, encodes an oligosaccharide transporter and facilitates rhizosphere competency in Metarhizium robertsii. Plant Physiol 154:1549–1557
Froehlich AC, Chen CH, Belden WJ, Madeti C, Roenneberg T, Merrow M, Loros JJ, Dunlap JC (2010) Genetic and molecular characterization of a cryptochrome from the filamentous fungus Neurospora crassa. Eukaryot Cell 9(5):738–750
Fuller KK, Dunlap JC, Loros JJ (2016) Fungal light sensing at the bench and beyond. Adv Genet 96:1–51
Gin E, Diernfellner ACR, Brunner M, Höfer T (2013) The Neurospora photoreceptor VIVID exerts negative and positive control on light sensing to achieve adaptation. Mol Syst Biol 9:667
Godin KS, Varani G (2007) How arginine-rich domains coordinate mRNA maturation events. RNA Biol 4(2):69–75
Guo MM, Guo SP, Yang HJ, Bu N, Dong CH (2016) Comparison of major bioactive compounds of the caterpillar medicinal mushroom, Cordyceps militaris (ascomycetes), fruiting bodies cultured on wheat substrate and pupae. Int J Med Mushrooms 18(4):327–336
Heintzen C, Loros JJ, Dunlap JC (2001) The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell 104(3):453–464
Hitomi K, Okamoto K, Daiysasu H, Miyashita H, Iwai S, Toh H, Ishiura M, Todo T (2000) Bacterial cryptochrome and photolyase: characterization of two photolyase-like genes of Synechocystis sp. PCC6803. Nucleic Acids Res 28(12):2352–2362
Huang Y, Baxter R, Smith BS, Partch CL, Colbert CL, Deisenhofer J (2006) Crystal structure of cryptochrome 3 from Arabidopsis thaliana and its implications for photolyase activity. Proc Natl Acad Sci U S A 103(47):17701–17706
Idnurm A, Heitman J (2005) Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol 3:e95
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780
Khang CH, Park SY, Rho HS, Lee YH, Kang S (2007) Filamentous fungi (Magnaporthe grisea and Fusarium oxysporum). In: Wang K (ed) Agrobacterium protocols, vol 2. Humana, Totowa, pp 403–420
Kornerup A, Wanscher JH (1978) Methuen handbook of colour. EyreMethuen, London
Lian TT, Yang T, Liu GJ, Sun JD, Dong CH (2014) Reliable reference gene selection for Cordyceps militaris gene expression studies under different developmental stages and media. FEMS Microbiol Lett 356(1):97–104
Linden H, Ballario P, Macino G (1997) Blue light regulation in Neurospora crassa. Fungal Genet Biol 22:141–150
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and 2-∆∆C(T) method. Methods 25:402–408
Mei QM, Dvornyk V (2015) Evolutionary history of the photolyase/cryptochrome superfamily in eukaryotes. PLoS One 10(9):e0135940
Mullins ED, Chen X, Romaine P, Raina R, Geiser DM, Kang S (2001) Agrobacterium-mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology 91(2):173–180
Nsa IY, Karunarathna N, Liu X, Huang H, Boetteger B, Bell-Pedersen D (2015) A novel cryptochrome-dependent oscillator in Neurospora crassa. Genetics 199(1):233–245
Olmedo M, Ruger-Herreros C, Luque EM, Corrochano LM (2010) A complex photoreceptor system mediates the regulation by light of the conidiation genes con-10 and con-6 in Neurospora crassa. Fungal Genet Biol 47(4):352–363
Pokorny R, Klar T, Hennecke U, Carell T, Batschauer A, Essen LO (2008) Recognition and repair of UV lesions in loop structures of duplex DNA by DASH-type cryptochrome. Proc Natl Acad Sci U S A 105(52):21023–21027
Sancar A (2003) Structure and function of DNA photolyase and cryptochrome blue light photoreceptors. Chem Rev 103(6):2203–2237
Sato H, Shimazu M (2002) Stromata production for Cordyceps militaris (Clavicipitales: Clavicipitaceae) by injection of hyphal bodies to alternative host insects. Appl Entomol Zoo l37:85–92
Scheerer P, Zhang F, Kalms J, von Stetten D, Krauβ N, Oberpichler I, Lamparter T (2015) The class III cyclobutane pyrimidine dimer photolyase structure reveals a new antenna chromophore binding site and alternative photoreduction pathways. J Biol Chem 290(18):11504–11514
Schwerdtfeger C, Linden H (2001) Blue light adaptation and desensitization of light signal transduction in Neurospora crassa. Mol Microbiol 39:1080–1087
Schwerdtfeger C, Linden H (2003) VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J 22(18):4846–4855
Selby CP, Sancar A (2006) A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc Natl Acad Sci U S A 103(47):17696–17700
Smith KM, Sancar G, Dekhang R, Stajich CM, Li SJ, Tag AG, Sancar C, Bredeweg EL, Priest HD, McCormick RF, Thomas TL, Carrington JC, Stajich JE, Bell-Pedersen D, Brunner M, Freitag M (2010) Transcription factors in light and circadian clock signaling networks revealed by genome wide mapping of direct targets for Neurospora white collar complex. Eukaryot Cell 9(10):1549–1556
Tagua VG, Pausch M, Eckel M, Gutiérrez G, Miralles-Durán A, Sanz C, Eslava AP, Pokorny R, Corrochano LM, Batschauer A (2015) Fungal cryptochrome with DNA repair activity reveals an early stage in cryptochrome evolution. Proc Natl Acad Sci U S A 112(49):15130–15135
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol and Evol 30(12):2725–2729
Thandapani P, O’Connor TR, Bailey TL, Richard S (2013) Defining the RGG/RG motif. Mol Cell 50(5):613–623
Veluchamy S, Rollins JA (2008) A CRY-DASH-type photolyase/cryptochrome from Sclerotinia sclerotiorum mediates minor UV-A-specific effects on development. Fungal Genet Biol 45(9):1265–1276
Yang T, Dong CH (2014) Photo morphogenesis and photo response of the blue-light receptor gene Cmwc-1 in different strains of Cordyceps militaris. FEMS Microbiol Lett 352(2):190–197
Yang L, Ukil L, Osmani A, Nahm F, Davies J, De Souza CP, Dou X, Perez-Balaguer A, Osmani SA (2004) Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot Cell 3(5):1359–1362
Yang T, Sun JD, Lian TT, Wang WZ, Dong CH (2014) Process optimization or extraction of carotenoids from Cordyceps militaris, an edible and medicinal fungus. Int J Med Mushrooms 16(2):125–135
Yang T, Guo M, Yang HJ, Guo SP, Dong CH (2016) The blue light receptor CmWC-1 mediates fruit body development and secondary metabolism in Cordyceps militaris. Appl Microbiol Biotechnol 100(2):743–755
Yu J, Hamari Z, Han K, Seo J, Reyes-Domínguez Y, Scazzocchio C (2004) Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41(11):973–981
Zhang A, Lu P, Dahl-Roshak AM, Paress PS, Kennedy S, Tkacz JS, An Z (2003) Efficient disruption of a polyketide synthase gene (pks1) required for melanin synthesis through Agrobacterium-mediated transformation of Glarea lozoyensis. Mol Gen Genom 268(5):645–655
Zhang F, Scheerer P, Oberpichler I, Lamparter T, Krauβ N (2013) Crystal structure of a prokaryotic (6-4) photolyase with an Fe-S cluster and a 6,7-dimethyl-8-ribityllumazine antenna chromophore. Proc Natl Acad Sci U S A 110(18):7217–7222
Zheng P, Xia YL, Xiao GH, Xiong CH, Hu X, Zhang SW, Zheng HY, Huang Y, Zhou Y, Wang SY, Zhao GP, Liu XZ, St Leger RJ, Wang CS (2011a) Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional Chinese medicine. Genome Biol 12(11):R116
Zheng Z, Huang C, Cao L, Xie C, Han R (2011b) Agrobacterium tumefaciens-mediated transformation as a tool for insertional mutagenesis in medicinal fungus Cordyceps militaris. Fungal Biol 115(3):265–274
Zheng ZL, Qiu XH, Han RC (2015) Identification of the genes involved in the fruiting body production and cordycepin formation of Cordyceps militaris. Mycobiology 43(1):37–42
Zhou X, Gong Z, Su Y, Lin J, Tang K (2009) Cordyceps fungi: natural products, pharmacological functions and developmental products. J Pharm Pharmacol 61(63):279–291
Acknowledgements
The authors are grateful to Prof. Xingzhong Liu and Wenbing Yin (Institute of Microbiology, Chinese Academy of Sciences) for providing the pAg1-H3 and pPk2-bar-gfp plasmids.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was funded by the National Natural Science Foundation of China (31572179, 31600054), by the Coal-based Key Scientific and Technological Project from Shanxi Province (FT2014-03-01), and the Key Research and Development Program from Guangxi Province (2016AB05317).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 183 kb)
Rights and permissions
About this article
Cite this article
Wang, F., Song, X., Dong, X. et al. DASH-type cryptochromes regulate fruiting body development and secondary metabolism differently than CmWC-1 in the fungus Cordyceps militaris . Appl Microbiol Biotechnol 101, 4645–4657 (2017). https://doi.org/10.1007/s00253-017-8276-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8276-7