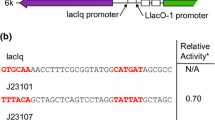
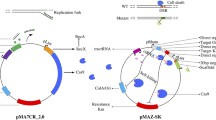
Abstract
Fine-tuning the expression level of multiple genes is usually pivotal for metabolic optimization. We have developed a tool for this purpose for the important industrial workhorse Corynebacterium glutamicum that allows for the introduction of synthetic promoter-driven expression libraries of arbitrary genes. We first devised a method for introducing genetic elements into the chromosome repeatedly, relying on site-specific recombinases and the vector pJS31 serving as the carrier. The pJS31 vector contains a synthetic cassette including a phage attachment site attP for integration, a bacterial attachment site attB for subsequent integration, a multiple cloning site, and two modified loxP sites to facilitate easy removal of undesirable vector elements. Meanwhile, we constructed a derivative of the wild-type strain ATCC 13032 carrying an attB site in its chromosome (JS34) and demonstrated that pJS31 readily could integrate into the attB site in this strain providing expression of the corresponding integrase. Subsequent expression of the Cre recombinase promoted recombination between the modified loxP sites, resulting in a strain only retaining the target insertions and an attB site. To simplify the procedure, non-replicating circular expression units for the phage integrase and the Cre recombinase were used. As a showcase, we used the tool to construct a battery of strains simultaneously expressing the two reporter genes, lacZ (encoding β-galactosidase) and gusA (encoding β-glucuronidase), to arbitrary levels. In principle, an unlimited number of genes, whether native, heterologous, or synthetic, can be introduced using the developed approach, and this should greatly facilitate metabolic optimization of this important platform organism.







Similar content being viewed by others
References
Albert H, Dale EC, Lee E, Ow DW (1995) Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J 7:649–659. doi:10.1016/0168-9525(95)90182-5
Aristidou A, Penttilä M (2000) Metabolic engineering applications to renewable resource utilization. Curr Opin Biotechnol 11:187–198. doi:10.1016/S0958-1669(00)00085-9
Barrett E, Stanton C, Zelder O, Fitzgerald G, Ross RP (2004) Heterologous expression of lactose- and galactose-utilizing pathways from lactic acid bacteria in Corynebacterium glutamicum for production of lysine in whey. Appl Environ Microbiol 70:2861–2866. doi:10.1128/AEM.70.5.2861-2866.2004
Becker J, Wittmann C (2012) Bio-based production of chemicals, materials and fuels—Corynebacterium glutamicum as versatile cell factory. Curr Opin Biotechnol 23:631–640. doi:10.1016/j.copbio.2011.11.012
Binder S, Siedler S, Marienhagen J, Bott M, Eggeling L (2013) Recombineering in Corynebacterium glutamicum combined with optical nanosensors: a general strategy for fast producer strain generation. Nucleic Acids Res 41(12):6360–6369. doi:10.1093/nar/gkt312
Breüner A, Brøndsted L, Hammer K (2001) Resolvase-like recombination performed by the TP901-1 integrase. Microbiology 147:2051–2063
Brøndsted L, Hammer K (1999) Use of the integration elements encoded by the temperate lactococcal bacteriophage TP901-1 to obtain chromosomal single-copy transcriptional fusions in Lactococcus lactis. Appl Environ Microbiol 65:752–758
Buchholz J, Schwentner A, Brunnenkan B, Gabris C, Grimm S, Gerstmeir R, Eikmanns BJ, Blombach B (2013) Platform engineering of Corynebacterium glutamicum with reduced pyruvate dehydrogenase complex activity for improved production of L-lysine, L-valine, and 2-ketoisovalerate. Appl Environ Microbiol 18:5566–5575. doi:10.1128/AEM.01741-13
Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kühn R (2015) Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol 33:543–548. doi:10.1038/nbt.3198
Cleto S, Jensen JVK, Wendisch VF, Lu TK (2016) ACS Synth Biol 5:375–385
Cohen N, April R (1980) Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Microbiol Biotechnol 138:179–207
Eggeling L, Bott M (2005) Handbook of Corynebacterium glutamicum. CRC, Boca Raton
Fell DA, Thomas S (1995) Physiological control of metabolic flux: the requirement for multisite modulation. Biochem J 311:35–39
Friehs K (2004) Plasmid copy number and plasmid stability. Adv Biochem Eng Biotechnol 86:47–82
Fukui K, Koseki C, Yamamoto Y, Nakamura J, Sasahara A, Yuji R, Hashiguchi K, Usuda Y, Matsui K, Kojima H, Abe K (2011) Identification of succinate exporter in Corynebacterium glutamicum and its physiological roles under anaerobic conditions. J Biotechnol 154:25–34. doi:10.1016/j.jbiotec.2011.03.010
Glick BR (1995) Metabolic load and heterologous gene expression. Biotechnol Adv 13:247–261
Hermann T (2003) Industrial production of amino acids by coryneform bacteria. J Biotechnol 104:155–172. doi:10.1016/S0168-1656(03)00149-4
Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278. doi:10.1016/j.cell.2014.05.010
Israelsen H, Madsen SM, Vrang A, Hansen EB, Johansen E (1995) Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector, pAK80. Appl Environ Microbiol 61:2540–2547. doi:10.1128/AEM.68.10.5051-5056.2002
Jäger W, Schäfer A, Pühler A, Labes G, Spring S (1992) Expression of the Bacillus subtilis sacB gene leads to sucrose sensitivity in the Gram-positive bacterium Corynebactenium glutamicum but not in Streptomyces lividans. J Bacteriol 174:5462–5465
Jensen PR, Westerhoff HV, Michelsen O (1993) The use of lac-type promoters in control analysis. Eur J Biochem 211:181–191. doi:10.1111/j.1432-1033.1993.tb19885.x
Kawaguchi H, Vertès AA, Okino S, Verte AA, Inui M, Yukawa H (2006) Engineering of a xylose metabolic pathway in Corynebacterium glutamicum. Appl Environ Microbiol 5:3418–3428. doi:10.1128/AEM.72.5.3418
Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory, New York
Moreau S, Blanco C, Trautwetter A (1995) Site-specific integration of corynephage phi16: construction of an integration vector. Microbiology 145:539–548
Neidhardt FC, Bloch PL, Smith DF (1974) Culture medium for Enterobacteria. J Bacteriol 119:736–747
Nešvera J, Pátek M (2011) Tools for genetic manipulations in Corynebacterium glutamicum and their applications. Appl Microbiol Biotechnol 90:1641–1654. doi:10.1007/s00253-011-3272-9
Nielsen HJ, Ottesen JR, Youngren B, Austin SJ, Hansen FG (2006) The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol Microbiol 62:331–338. doi:10.1111/j.1365-2958.2006.05346.x
Novick RP (1987) Plasmid incompatibility. Microbiol Rev 51:381–395. doi:10.1016/0147-619X(78)90001-X
Pátek M, Holátko J, Busche T, Kalinowski J, Nešvera J (2013) Corynebacterium glutamicum promoters: a practical approach. Microb Biotechnol 6:103–117. doi:10.1111/1751-7915.12019
Petersen KV, Martinussen J, Jensen PR, Solem C (2013) Repetitive, marker-free, site-specific integration as a novel tool for multiple chromosomal integration of DNA. Appl Environ Microbiol 79:3563–3569. doi:10.1128/AEM.00346-13
Pfleger BF, Pitera DJ, Smolke CD, Keasling JD (2006) Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat Biotechnol 24:1027–1032
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. doi:10.1038/nprot.2013.143
Rytter JV, Helmark S, Chen J, Lezyk MJ, Solem C, Jensen PR (2014) Synthetic promoter libraries for Corynebacterium glutamicum. Appl Microbiol Biotechnol 98:2617. doi:10.1007/s00253-013-5481-x
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, New York
Sander JD, Joung JK (2014) CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32:347–355. doi:10.1038/nbt.2842
Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73
Selbitschka W, Niemann S, Piihler A (1993) Construction of gene replacement vectors for Gram bacteria using a genetically modified sacRB gene as a positive selection marker. Appl Microbiol Biotechnol 38:615–618. doi:10.1007/BF00182799
Shukuo K, Shigezo U, Masakazu S (1957) Studies on the amino acid fermentation. J Gen Appl Microbiol 3:193–205. doi:10.2323/jgam.3.193
Solem C, Koebmann BJ, Jensen PR (2003) Glyceraldehyde-3-phosphate dehydrogenase has no control over glycolytic flux in Lactococcus lactis MG1363. J Bacteriol 185:1564–1571. doi:10.1128/JB.185.5.1564-1571.2003
Stoll SM, Stoll SM, Ginsburg DS, Ginsburg DS, Calos MP, Calos MP (2002) Phage TP901-1 site-specifc integrase functions in human cells. J Bacteriol 184:3657–3663. doi:10.1128/JB.184.13.3657
Suzuki N, Inui M, Yukawa H (2007) Site-directed integration system using a combination of mutant lox sites for Corynebacterium glutamicum. Appl Microbiol Biotechnol 77:871–878. doi:10.1007/s00253–007–1215-2
Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R (2013) One-step generation of mice carrying reporter and conditional alleles by CRISPR/cas-mediated genome engineering. Cell 154:1370–1379. doi:10.1016/j.cell.2013.08.022
Acknowledgments
This work was supported by grant NNF12OC0000818 from the Novo Nordisk Foundation and by the Bio-Value Strategic Platform for Innovation and Research which is co-funded by the Danish Council for Strategic Research and the Danish Council for Technology and Innovation, grant no.: 0603-00522B.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The article does not contain any studies with animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 382 kb)
Rights and permissions
About this article
Cite this article
Shen, J., Chen, J., Jensen, P.R. et al. A novel genetic tool for metabolic optimization of Corynebacterium glutamicum: efficient and repetitive chromosomal integration of synthetic promoter-driven expression libraries. Appl Microbiol Biotechnol 101, 4737–4746 (2017). https://doi.org/10.1007/s00253-017-8222-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8222-8