Abstract
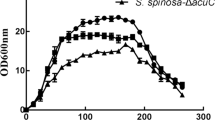
Saccharopolyspora spinosa can produce spinosad as a major secondary metabolite, which is an environmentally friendly agent for insect control. Cobalamin-independent methionine synthase (MetE) is an important enzyme in methionine biosynthesis, and this enzyme is probably closely related to spinosad production. In this study, its corresponding gene metE was inactivated, which resulted in a rapid growth and glucose utilisation rate and almost loss of spinosad production. A label-free quantitative proteomics-based approach was employed to obtain insights into the mechanism by which the metabolic network adapts to the absence of MetE. A total of 1440 proteins were detected from wild-type and ΔmetE mutant strains at three time points: stationary phase of ΔmetE mutant strain (S1ΔmetE , 67 h), first stationary phase of wild-type strain (S1WT, 67 h) and second stationary phase of wild-type strain (S2WT, 100 h). Protein expression patterns were determined using an exponentially modified protein abundance index (emPAI) and analysed by comparing S1ΔmetE /S1WT and S1ΔmetE /S2WT. Results showed that differentially expressed enzymes were mainly involved in primary metabolism and genetic information processing. This study demonstrated that the role of MetE is not restricted to methionine biosynthesis but rather is involved in global metabolic regulation in S. spinosa.




Similar content being viewed by others
References
Bierman M, Logan R, O’Brien K, Seno ET, Rao RN, Schoner BE (1992) Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49
Bologna FP, Andreo CS, Drincovich MF (2007) Escherichia coli malic enzymes: two isoforms with substantial differences in kinetic properties, metabolic regulation, and structure. J Bacteriol 189:5937–5946. doi:10.1128/JB.00428-07
Borodina I, Schöller C, Eliasson A, Nielsen J (2005) Metabolic network analysis of Streptomyces tenebrarius, a Streptomyces species with an active Entner-Doudoroff pathway. Appl Environ Microbiol 71:2294–2302. doi:10.1128/AEM.71.5.2294-2302.2005
Chopra T, Hamelin R, Armand F, Chiappe D, Moniatte M, McKinney JD (2014) Quantitative mass spectrometry reveals plasticity of metabolic networks in Mycobacterium smegmatis. Mol Cell Proteomics 13:3014–3028. doi:10.1074/mcp.M113.034082
Commichau FM, Gunka K, Landmann JJ, Stülke J (2008) Glutamate metabolism in Bacillus subtilis: gene expression and enzyme activities evolved to avoid futile cycles and to allow rapid responses to perturbations of the system. J Bacteriol 190:3557–3564. doi:10.1128/JB.00099-08
Gallo G, Alduina R, Renzone G, Thykaer J, Bianco L, Eliasson-Lantz A, Scaloni A, Puglia AM (2010) Differential proteomic analysis highlights metabolic strategies associated with balhimycin production in Amycolatopsis balhimycina chemostat cultivations. Microb Cell Factories 9:95. doi:10.1186/1475-2859-9-95
Gallo G, Piccolo LL, Renzone G, La Rosa R, Scaloni A, Quatrini P, Puglia AM (2012) Differential proteomic analysis of an engineered Streptomyces coelicolor strain reveals metabolic pathways supporting growth on n-hexadecane. Appl Microbiol Biotechnol 94:1289–1301. doi:10.1007/s00253-012-4046-8
Görisch H (2003) The ethanol oxidation system and its regulation in Pseudomonas aeruginosa. BBA-Proteins Proteom 1647:98–102. doi:10.1016/S1570-9639(03)00066-9
Grossmann J, Roschitzki B, Panse C, Fortes C, Barkow-Oesterreicher S, Rutishauser D, Schlapbach R (2010) Implementation and evaluation of relative and absolute quantification in shotgun proteomics with label-free methods. J Proteomics 73:1740–1746. doi:10.1016/j.jprot.2010.05.011
Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580
Henard CA, Bourret TJ, Song M, Vázquez-Torres A (2010) Control of redox balance by the stringent response regulatory protein promotes antioxidant defenses of Salmonella. J Biol Chem 285:36785–36793. doi:10.1074/jbc.M110.160960
Huang S, Ding X, Sun Y, Yang Q, Xiao X, Cao Z, Xia L (2012) Proteomic analysis of Bacillus thuringiensis at different growth phases by using an automated online two-dimensional liquid chromatography-tandem mass spectrometry strategy. Appl Environ Microbiol 78:5270–5279. doi:10.1128/AEM.00424-12
Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4:1265–1272. doi:10.1074/mcp.M500061-MCP200
Jantama K, Zhang X, Moore JC, Shanmugam KT, Svoronos SA, Ingram LO (2008) Eliminating side products and increasing succinate yields in engineered strains of Escherichia coli C. Biotechnol Bioeng 101:881–893. doi:10.1002/bit.22005
Karp NA, McCormick PS, Russell MR, Lilley KS (2007) Experimental and statistical considerations to avoid false conclusions in proteomics studies using differential in-gel electrophoresis. Mol Cell Proteomics 6:1354–1364. doi:10.1074/mcp.M600274-MCP200
Krömer JO, Heinzle E, Schröder H, Wittmann C (2006) Accumulation of homolanthionine and activation of a novel pathway for isoleucine biosynthesis in Corynebacterium glutamicum McbR deletion strains. J Bacteriol 188:609–618. doi:10.1128/JB.188.2.609-618.2006
Kusch H, Engelmann S, Bode R, Albrecht D, Morschhäuser J, Hecker M (2008) A proteomic view of Candida albicans yeast cell metabolism in exponential and stationary growth phases. Int J Med Microbiol 298:291–318. doi:10.1016/j.ijmm.2007.03.020
Lee EG, Yoon SH, Das A, Lee SH, Li C, Kim JY, Choi MS, Oh DK, Kim WS (2009) Directing vanillin production from ferulic acid by increased acetyl‐CoA consumption in recombinant Escherichia coli. Biotechnol Bioeng 102:200–208. doi:10.1002/bit.22040
Li L, Wada M, Yokota A (2007) A comparative proteomic approach to understand the adaptations of an H+‐ATPase‐defective mutant of Corynebacterium glutamicum ATCC14067 to energy deficiencies. Proteomics 7:3348–3357. doi:10.1002/pmic.200700287
Li S, Ha SJ, Kim HJ, Galazka JM, Cate JH, Jin YS, Zhao H (2013) Investigation of the functional role of aldose 1-epimerase in engineered cellobiose utilization. J Biotechnol 168:1–6. doi:10.1016/j.jbiotec.2013.08.003
Licona-Cassani C, Lim S, Marcellin E, Nielsen LK (2014) Temporal dynamics of the Saccharopolyspora erythraea phosphoproteome. Mol Cell Proteomics 13:1219–1230. doi:10.1074/mcp.M113.033951
Liu F, Hao J, Yan H, Bach T, Fan L (2014) AspC-mediated aspartate metabolism coordinates the Escherichia coli cell cycle. PLoS ONE 9, e92229. doi:10.1371/journal.pone.0092229
Lu D, Liu RZ, Izumi V, Fenstermacher D, Haura EB, Koomen J, Eschrich SA (2008) IPEP: an in silico tool to examine proteolytic peptides for mass spectrometry. Bioinformatics 24:2801–2802. doi:10.1093/bioinformatics/btn511
Luo Y, Ding X, Xia L, Huang F, Li W, Huang S, Tang Y, Sun Y (2011) Comparative proteomic analysis of Saccharopolyspora spinosa SP06081 and PR2 strains reveals the differentially expressed proteins correlated with the increase of spinosad yield. Proteome Sci 9:1–12. doi:10.1186/1477-5956-9-40
Manteca A, Sanchez J, Jung HR, Schwämmle V, Jensen ON (2010) Quantitative proteomics analysis of Streptomyces coelicolor development demonstrates that onset of secondary metabolism coincides with hypha differentiation. Mol Cell Proteomics 9:1423–1436. doi:10.1074/mcp.M900449-MCP200
Masuda T, Saito N, Tomita M, Ishihama Y (2009) Unbiased quantitation of Escherichia coli membrane proteome using phase transfer surfactants. Mol Cell Proteomics 8:2770–2777. doi:10.1074/mcp.M900240-MCP200
Michta E, Ding W, Zhu S, Blin K, Ruan H, Wang R, Wohlleben W, Mast Y (2014) Proteomic approach to reveal the regulatory function of aconitase AcnA in oxidative stress response in the antibiotic producer Streptomyces viridochromogenes Tü494. PLoS ONE 9, e87905. doi:10.1371/journal.pone.0087905
Mordukhova EA, Pan JG (2013) Evolved cobalamin-independent methionine synthase (MetE) improves the acetate and thermal tolerance of Escherichia coli. Appl Environ Microbiol 79:7905–7915. doi:10.1128/AEM.01952-13
Neilson KA, Ali NA, Muralidharan S, Mirzaei M, Mariani M, Assadourian G, Lee A, Sluyter SCV, Haynes PA (2011) Less label, more free: approaches in label‐free quantitative mass spectrometry. Proteomics 11:535–553. doi:10.1002/pmic.201000553
Novichkov PS, Li X, Kuehl JV, Deutschbauer AM, Arkin AP, Price MN, Rodionov DA (2014) Control of methionine metabolism by the SahR transcriptional regulator in Proteobacteria. Environ Microbiol 16:1–8. doi:10.1111/1462-2920.12273
Oh MK, Rohlin L, Kao KC, Liao JC (2002) Global expression profiling of acetate-grown Escherichia coli. J Biol Chem 277:13175–13183. doi:10.1074/jbc.M110809200
Okamoto S, Lezhava A, Hosaka T, Okamoto-Hosoya Y, Ochi K (2003) Enhanced expression of S-adenosylmethionine synthetase causes overproduction of actinorhodin in Streptomyces coelicolor A3 (2). J Bacteriol 185:601–609. doi:10.1128/JB.185.2.601-609.2003
Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1:376–386. doi:10.1074/mcp.M200025-MCP200
Pejchal R, Ludwig ML (2004) Cobalamin-independent methionine synthase (MetE): a face-to-face double barrel that evolved by gene duplication. PLoS Biol 3, e31. doi:10.1371/journal.pbio.0030031
Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP (2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res 2:43–50. doi:10.1021/pr025556v
Rappsilber J, Ryder U, Lamond AI, Mann M (2002) Large-scale proteomic analysis of the human spliceosome. Genome Res 12:1231–1245. doi:10.1101/gr.473902
Schreier HJ, Dejtisakdi W, Escalante JO, Brailo M (2012) Transposon mutagenesis of Planctomyces limnophilus and analysis of a pckA mutant. Appl Environ Microbiol 78:7120–7123. doi:10.1128/AEM.01794-12
Simon R, Priefer U, Pühler A (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat Biotechnol 1:784–791. doi:10.1038/nbt1183-784
Suda M, Teramoto H, Imamiya T, Inui M, Yukawa H (2008) Transcriptional regulation of Corynebacterium glutamicum methionine biosynthesis genes in response to methionine supplementation under oxygen deprivation. Appl Microbiol Biotechnol 81:505–513. doi:10.1007/s00253-008-1694-9
Takano H, Hagiwara K, Ueda K (2015) Fundamental role of cobalamin biosynthesis in the developmental growth of Streptomyces coelicolor A3 (2). Appl Microbiol Biotechnol 99:2329–2337. doi:10.1007/s00253-014-6325-z
Tang Y, Xia L, Ding X, Luo Y, Huang F, Jiang Y (2011) Duplication of partial spinosyn biosynthetic gene cluster in Saccharopolyspora spinosa enhances spinosyn production. FEMS Microbiol Lett 325:22–29. doi:10.1111/j.1574-6968.2011.02405.x
Thomas MG, Chan YA, Ozanick SG (2003) Deciphering tuberactinomycin biosynthesis: isolation, sequencing, and annotation of the viomycin biosynthetic gene cluster. Antimicrob Agents Chemother 47:2823–2830. doi:10.1128/AAC.47.9.2823-2830.2003
Thomas L, Hodgson DA, Wentzel A, Nieselt K, Ellingsen TE, Moore J, Morrissey ER, Legaie R, The STREAM Consortium, Wohlleben W, Rodríguez-García A, Martín JF, Burroughs NJ, Wellington EMH, Smith MCM (2012) Metabolic switches and adaptations deduced from the proteomes of Streptomyces coelicolor wild type and phoP mutant grown in batch culture. Mol Cell Proteomics 11:M111–M013797. doi:10.1074/mcp.M111.013797
Thompson GD, Dutton R, Sparks TC (2000) Spinosad—a case study: an example from a natural products discovery programme. Pest Manag Sci 56:696–702
Tiffert Y, Franz-Wachtel M, Fladerer C, Nordheim A, Reuther J, Wohlleben W, Mast Y (2011) Proteomic analysis of the GlnR-mediated response to nitrogen limitation in Streptomyces coelicolor M145. Appl Microbiol Biotechnol 89:1149–1159. doi:10.1007/s00253-011-3086-9
Wang R, Marcotte EM (2008) The proteomic response of Mycobacterium smegmatis to anti-tuberculosis drugs suggests targeted pathways. J Proteome Res 7:855–865. doi:10.1021/pr0703066
Wang Y, Wu SL, Hancock WS, Trala R, Kessler M, Taylor AH, Patel PS, Aon JC (2005) Proteomic profiling of Escherichia coli proteins under high cell density fed-batch cultivation with overexpression of phosphogluconolactonase. Biotechnol Prog 21:1401–1411. doi:10.1021/bp050048m
Wang Y, Wang Y, Chu J, Zhuang Y, Zhang L, Zhang S (2007) Improved production of erythromycin A by expression of a heterologous gene encoding S-adenosylmethionine synthetase. Appl Microbiol Biotechnol 75:837–842. doi:10.1007/s00253-007-0894-z
Wick LM, Quadroni M, Egli T (2001) Short‐and long‐term changes in proteome composition and kinetic properties in a culture of Escherichia coli during transition from glucose‐excess to glucose‐limited growth conditions in continuous culture and vice versa. Environ Microbiol 3:588–599
Wiese S, Reidegeld KA, Meyer HE, Warscheid B (2007) Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics 7:340–350. doi:10.1002/pmic.200600422
Wilkins MJ, Callister SJ, Miletto M, Williams KH, Nicora CD, Lovley DR, Long PE, Lipton MS (2011) Development of a biomarker for Geobacter activity and strain composition; proteogenomic analysis of the citrate synthase protein during bioremediation of U (VI). Microb Biotechnol 4:55–63. doi:10.1111/j.1751-7915.2010.00194.x
Yang Q, Ding X, Liu X, Liu S, Sun Y, Yu Z, Hu S, Rang J, He H, He L, Xia L (2014) Differential proteomic profiling reveals regulatory proteins and novel links between primary metabolism and spinosad production in Saccharopolyspora spinosa. Microb Cell Factories 13:27. doi:10.1186/1475-2859-13-27
Yang Q, Tang S, Rang J, Zuo M, Ding X, Sun Y, Feng P, Xia L (2015) Detection of toxin proteins from Bacillus thuringiensis strain 4.0718 by strategy of 2D-LC–MS/MS. Curr Microbiol 70:457–463. doi:10.1007/s00284-014-0747-9
Ying J, Wang H, Bao B, Zhang Y, Zhang J, Zhang C, Li A, Lu J, Li P, Ying J, Liu Q, Xu T, Yi H, Li J, Zhou L, Zhou T, Xu Z, Ni L, Bao Q (2015) Molecular variation and horizontal gene transfer of the homocysteine methyltransferase gene mmuM and its distribution in clinical pathogens. Int J Biol Sci 11:11. doi:10.7150/ijbs.10320
Zhao XQ, Gust B, Heide L (2010) S-adenosylmethionine (SAM) and antibiotic biosynthesis: effect of external addition of SAM and of overexpression of SAM biosynthesis genes on novobiocin production in Streptomyces. Arch Microbiol 192:289–297. doi:10.1007/s00203-010-0548-x
Acknowledgments
This work was supported by the National Basic Research Program (973) of China (2012CB722301), the National High Technology Research and Development program (863) of China (2011AA10A203), the International Cooperation Project (0102011DFA32610) and the Cooperative Innovation Center of Engineering and New Products for Developmental Biology of Hunan Province (20134486).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Qi Yang and Yunlong Li contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 2237 kb)
Rights and permissions
About this article
Cite this article
Yang, Q., Li, Y., Yang, H. et al. Proteomic insights into metabolic adaptation to deletion of metE in Saccharopolyspora spinosa . Appl Microbiol Biotechnol 99, 8629–8641 (2015). https://doi.org/10.1007/s00253-015-6883-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6883-8